Figure 6.
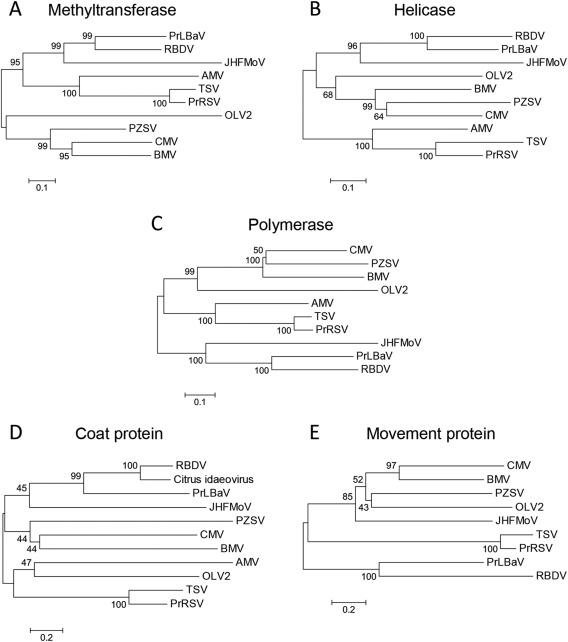
Phylogenetic trees inferred by neighbour‐joining analysis of the methyltransferase (A), helicase (B) and RNA‐dependent RNA polymerase (C) signatures, and of the coat proteins (D) and movement proteins (E) of privet leaf blotch‐associated virus (PrLBaV), RBDV (genus Idaeovirus), JHFMoV and representative viruses in the family Bromoviridae. The number on each branch is the result of bootstrap analysis (1000 replicates). Extended names of the viruses included in the phylogenetic trees are reported below, with the accession numbers of the respective methyltransferase (MTR), helicase (HEL), RNA‐dependent RNA polymerase (RdRp) signatures, and of the coat protein (CP) and movement protein (MP) reported in parentheses. AMV (Alfalfa mosaic virus; MTR and HEL, NP_041192; RdRp, YP_053235; CP, NP_041195; MP, NP_041194), BMV (Brome mosaic virus; MTR and HEL, NP_041196; RdRp, NP_041197; CP, AAA46334; MP, NP_041198), CMV (Cucumber mosaic virus; MTR and HEL, NP_049323; RdRp, NP_049324; CP, NP_040777; MP, NP_040776), JHMoV (Japanese holly fern mottle virus; MTR, HEL and RdRp, YP_003126903; CP, ACT67468; MP, YP_003126905), OLV2 (Olive latent virus 2; MTR and HEL, NP_620042; RdRp, NP_620043; CP, NP_620039; MP, NP_620038), PrRSV (Privet ringspot virus; MTR and HEL, YP_009165996; RdRp, YP_009165997; CP, YP_009166000; MP, YP_009165999), PZSV (Pelargonium zonate spot virus; MTR and HEL, NP_619770; RdRp, NP_619771; CP, NP_619773; MP, NP_619772), RBDV (Raspberry bushy dwarf virus; MTR, HEL and RdRp, NP_620465; CP, NP_620467; MP, NP_620466), TSV (Tobacco streak virus; MTR and HEL, NP_620772; RdRp, NP_620768; CP, NP_620774; MP, NP_620773), Citrus idaeovirus (CP; DQ100358).
