Summary
Citrus canker, caused by Xanthomonas citri, affects most commercial citrus varieties. All X. citri strains possess at least one transcription activator‐like effector of the PthA family that activates host disease susceptibility (S) genes. The X. citri strain 306 encodes four PthA effectors; nevertheless, only PthA4 is known to elicit cankers on citrus. As none of the PthAs act as avirulence factors on citrus, we hypothesized that PthAs 1–3 might also contribute to pathogenicity on certain hosts. Here, we show that, although PthA4 is indispensable for canker formation in six Brazilian citrus varieties, PthAs 1 and 3 contribute to canker development in ‘Pera’ sweet orange, but not in ‘Tahiti’ lemon. Deletions in two or more pthA genes reduce bacterial growth in planta more pronouncedly than single deletions, suggesting an additive role of PthAs in pathogenicity and bacterial fitness. The contribution of PthAs 1 and 3 in canker formation in ‘Pera’ plants does not correlate with the activation of the canker S gene, LOB1 (LATERAL ORGAN BOUNDARIES 1), but with the induction of other PthA targets, including LOB2 and citrus dioxygenase (DIOX). LOB1, LOB2 and DIOX show differential PthA‐dependent expression between ‘Pera’ and ‘Tahiti’ plants that appears to be associated with nucleotide polymorphisms found at or near PthA‐binding sites. We also present evidence that LOB1 activation alone is not sufficient to elicit cankers on citrus, and that DIOX acts as a canker S gene in ‘Pera’, but not ‘Tahiti’, plants. Our results suggest that the activation of multiple S genes, such as LOB1 and DIOX, is necessary for full canker development.
Keywords: citrus canker, citrus dioxygenase, LATERAL ORGAN BOUNDARIES genes, pthA, TAL effectors, Xanthomonas citri, Xanthomonas aurantifolii
Introduction
Citrus canker, caused by strains of Xanthomonas citri (also known as X. citri ssp. citri), is one of the most economically important citrus diseases that not only affects the commercial citrus plantations, but also the citrus markets worldwide (Brunings and Gabriel, 2003; Graham et al., 2004). The X. citri ‘A’ strains (Asiatic group) cause the most severe types of canker and have a broad host range, affecting sweet oranges (Citrus sinensis), grapefruits (Citrus paradisi) and lemons (Citrus limon). However, the citrus canker pathogens Xanthomonas aurantifolii (also known as X. fuscans ssp. aurantifolii) pathotypes ‘B’ and ‘C’ have a much narrower range of citrus hosts and are responsible for the ‘B’ and ‘C’ types of canker, respectively, which are restricted to some regions of South America (Brunings and Gabriel, 2003; Shubert et al., 2003). In particular, a ‘C’ strain of X. aurantifolii, isolated in Brazil, infects ‘Mexican’ lime only (Citrus aurantifolia) and induces a hypersensitive response (HR) in several citrus hosts, including sweet oranges and lemons (Brunings and Gabriel, 2003; Cernadas et al., 2008; Chiesa et al., 2013).
The disease symptoms caused by X. citri and X. aurantifolii ‘C’ are characterized by water‐soaked and eruptive lesions on the surface of leaves, twigs and fruits (Brunings and Gabriel, 2003; Cernadas et al., 2008; Shubert et al., 2003). The vigorous growth of the host cells, as a result of infection, causes the epidermis to break open, favouring pathogen spread and disease dissemination (Brunings and Gabriel, 2003; Wichmann and Bergelson, 2004). Although the precise mechanism by which these Xanthomonas pathogens induce canker is unknown, it is well known that members of the PthA/AvrBs3 family of transcriptional activator‐like (TAL) effectors play a central role in the activation of host genes implicated in cell division and growth (Al‐Saadi et al., 2007; Duan et al., 1999; Hu et al., 2014; Pereira et al., 2014; Soprano et al., 2013; Swarup et al., 1992; Yan and Wang, 2012).
TAL effectors function as transcription factors in plant cells. These proteins have a variable DNA‐binding domain made up of tandem repeats of 33–34 amino acids that recognizes specific promoter elements of host target genes (Boch and Bonas, 2010). The DNA‐binding specificity of TAL effectors is provided primarily by the highly polymorphic residues located at positions 12–13 of the repeats, known as repeat variable diresidues, or RVDs (Boch et al., 2009; Moscou and Bogdanove, 2009). Structural studies have revealed that the DNA‐binding domain of TAL effectors folds into a super‐helical structure that wraps around the DNA, and that the 13th RVD residue of each repeat makes direct contact with one DNA base in a linear fashion (Deng et al., 2012; Mak et al., 2012; Murakami et al., 2010). Therefore, as distinct RVDs show preferential binding to certain DNA bases, one can predict the DNA‐binding sequence or effector‐binding element (EBE) of a TAL effector by knowing its RVD composition (Boch et al., 2009; Moscou and Bogdanove, 2009).
TAL effectors play critical roles in disease development, acting as major pathogenicity determinants; however, they can also act as avirulence factors through the activation of defence‐related genes (Antony et al., 2010; Kay et al., 2007; Römer et al., 2007; Yang et al., 2006; Yu et al., 2011). Although some PthA variants have been suggested to act as avirulence factors on citrus (Chiesa et al., 2013; Shiotani et al., 2007), PthAs are generally thought to function as pathogenicity determinants that transactivate the host genes required for disease susceptibility and canker formation (Al‐Saadi et al., 2007; Cernadas et al., 2008; Hu et al., 2014; Pereira et al., 2014).
The citrus LATERAL ORGAN BOUNDARIES 1 (LOB1) gene is, to our knowledge, the only citrus canker susceptibility (S) gene known to date (Hu et al., 2014; Li et al., 2014; Pereira et al., 2014). It was found that certain PthA variants bind specifically to a region of the citrus LOB1 gene promoter enhancing LOB1 expression, and that the PthA‐dependent expression of LOB1 correlates with canker symptom development (Hu et al., 2014). In addition to LOB1, however, several citrus genes implicated in cell division, cell wall remodelling and auxin and gibberellin synthesis and action, including LOB2, LOB3 and DIOX (citrus dioxygenase), have been identified as potential direct targets of PthAs (Pereira et al., 2014). Nevertheless, the functional role played by these genes in canker elicitation and development remains to be elucidated.
All strains of X. citri and X. aurantifolii causing hyperplastic lesions on citrus carry at least one PthA variant that activates host S genes, including LOB1 (Al‐Saadi et al., 2007; Hu et al., 2014). For instance, the X. citri ‘A’ strains 306 and 3213 contain four pthA genes that encode highly homologous PthA proteins that differ from each other primarily in their DNA‐binding domain. Nevertheless, pathogenicity studies have shown that only pthA4 is necessary and sufficient for canker elicitation on ‘Valencia’ sweet orange and ‘Duncan’ grapefruit (Al‐Saadi et al., 2007; Hu et al., 2014; da Silva et al., 2002; Yan and Wang, 2012). In addition, none of the PthA proteins from these strains appear to act as avirulence factors on citrus plants (Al‐Saadi et al., 2007; Yan and Wang, 2012). Therefore, the existence of multiple pthA genes in a single bacterial strain suggests that each might contribute to disease development or pathogen fitness on certain hosts. The observation that PthAs form homo‐ and heterodimers in yeast cells, and that the transient expression of PthA2 or PthA4 in citrus epicotyls induces a similar category of functionally related genes associated with cell division and growth, supports this idea (Domingues et al., 2010; Pereira et al., 2014). Moreover, in spite of the differences in their RVD composition, transiently expressed PthAs 2 and 4 activate many common targets in citrus epicotyls (Pereira et al., 2014).
Here, we present evidence indicating that PthAs from X. citri strain 306 have additive roles in canker development and bacterial growth in some citrus hosts, including ‘Pera’ sweet orange. In addition to PthA4, PthAs 1 and 3 contribute to symptom development in ‘Pera’ leaves and this correlates with a PthA‐dependent activation of citrus DIOX, but not LOB1. Importantly, we show that, although LOB1 is regarded as an important citrus canker S gene (Hu et al., 2014; Li et al., 2014; Pereira et al., 2014), its induction is not always accompanied by canker formation, indicating that the activation of additional PthA targets is needed for full disease symptoms. In line with this idea, we present evidence suggesting that citrus DIOX, which encodes a member of the 2‐oxoglutarate/Fe(II)‐dependent dioxygenase (2OGD) family, also functions as a canker S gene, as psoralen, an inhibitor of this class of enzymes, inhibits canker formation. Furthermore, we found many nucleotide polymorphisms in the citrus LOB1, LOB2 and DIOX promoters that overlap with PthA sites, which might explain the differential PthA‐dependent expression of these genes in distinct citrus hosts.
Results
Host‐dependent contribution of PthAs 1 and 3 to canker formation
Six commercial citrus varieties largely cultivated in Brazil, including the sweet oranges ‘Hamlin’, ‘Valencia’, ‘Pera’, ‘Sorocaba’ and ‘Natal’ (Washington Navel), and the ‘Tahiti’ lemon cultivar, were infiltrated with the single (Δ1, Δ3, Δ4), double (Δ1‐3, Δ1‐4, Δ3‐4) and triple (Δ1‐3‐4) pthA‐deletion mutants, and the canker symptoms were compared with those caused by the wild‐type bacterium (Figs 1 and 2). In agreement with previous studies (Al‐Saadi et al., 2007; Hu et al., 2014; Soprano et al., 2013; Yan and Wang, 2012), we found that pthA4 is indispensable for canker elicitation, as none of the citrus hosts inoculated with the Δ4 mutant, or with any of the mutants lacking pthA4, showed hyperplastic lesions on leaves (Figs 1 and 2). However, a deletion in pthA1 also caused a substantial reduction in the number of canker pustules in all the sweet orange varieties, particularly in ‘Hamlin’ and ‘Pera’, but not in ‘Tahiti’ lemon (Fig. 1). Similarly, a deletion in pthA3 reduced the severity of the canker lesions in ‘Pera’ and, to a lesser extent, in ‘Hamlin’ and ‘Sorocaba’, but not in ‘Valencia’, ‘Natal’ or ‘Tahiti’ plants (Fig. 1). Canker lesions were further reduced in most of the hosts inoculated with the Δ1‐3 mutant, suggesting that pthAs 1 and 3 have additive effects on pustule formation elicited by pthA4 (Fig. 2). The fact that the Δ1‐3‐4 mutant did not induce any symptoms in any of the citrus plants tested also suggests that pthA2 alone is not sufficient to elicit canker on citrus (Fig. 2). Together, these data show that PthAs 1 and 3 contribute to symptom development elicited by PthA4 in certain citrus hosts.
Figure 1.

Host‐dependent effect of single pthA deletions in canker development. Leaves of the sweet orange varieties ‘Hamlin’, ‘Valencia’, ‘Natal’, ‘Pera’ and ‘Sorocaba’, and ‘Tahiti’ lemon, were infiltrated with bacterial suspensions (105 cells/mL) of wild‐type (Wt) Xanthomonas citri and respective single pthA‐deletion mutants (Δ1, Δ3, Δ4). Canker symptoms were evaluated 14 days after bacterial inoculation. pthA4 is essential to elicit cankers in all citrus hosts; nevertheless, pthA1 also contributes significantly to symptom development in ‘Hamlin’, ‘Valencia’ and ‘Pera’, but not in ‘Tahiti’ plants. Similarly, a deletion in pthA3 also reduces canker formation in ‘Pera’ and, to a lesser extent, in ‘Hamlin’ and ‘Sorocaba’, but not in ‘Valencia’, ‘Natal’ or ‘Tahiti’ plants.
Figure 2.

Host‐dependent effect of double and triple pthA deletions in canker development. Leaves of ‘Hamlin’, ‘Valencia’, ‘Natal’, ‘Pera’, ‘Sorocaba’ and ‘Tahiti’ plants were infiltrated with bacterial suspensions (105 cells/mL) of wild‐type (Wt) Xanthomonas citri and respective double (Δ1‐3, Δ1‐4, Δ3‐4) and triple (Δ1‐3‐4) pthA‐deletion mutants, and canker symptoms were evaluated 14 days after bacterial inoculation. Although a deletion in pthA4 is sufficient to abolish cankers in all the citrus hosts, an additive effect of pthAs 1 and 3 in disease development is noted in ‘Pera’, ‘Natal’ and ‘Sorocaba’ plants.
PthAs affect bacterial growth in planta in an additive manner
The observation that PthAs 1 and 3 contribute to canker formation, particularly in ‘Pera’ sweet orange, but not ‘Tahiti’ lemon, led us to investigate whether they would also affect bacterial growth in planta. To test this, we infiltrated leaves of ‘Pera’ and ‘Tahiti’ plants with the pthA mutants and followed their growth compared with that of the wild‐type bacterium (Fig. 3). We found that all the pthA mutations reduced bacterial growth in both ‘Pera’ and ‘Tahiti’ leaves at 2 and 14 days post‐inoculation (dpi) relative to the wild‐type bacterium (Fig. 3). In addition, the double (Δ1‐4, Δ3‐4) and triple (Δ1‐3‐4) pthA‐deletion mutants grew significantly less well than the respective single mutants or the wild‐type bacterium in these hosts (Fig. 3). Similar results were observed in ‘Valencia’ and ‘Sorocaba’ plants (Fig. 3), indicating that PthAs 1, 3 and 4 exert an additive effect on bacterial growth in planta. Interestingly, however, although there seems to be a correlation between bacterial growth and the induction of cell hypertrophy, the Δ4 mutant reached bacterial titres higher than 108 cells/cm2 of leaf at 14 dpi in most of the hosts without causing any canker symptoms (Figs 1 and 3). In agreement with these results, Yan and Wang (2012) also reported that a pthA4 knockout mutant, which only caused chlorosis in grapefruit leaves, reached almost 108 bacterial cells/cm2 of leaf at 4 dpi.
Figure 3.
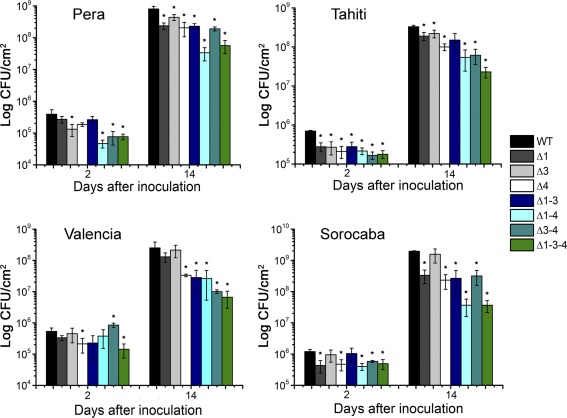
Additive effect of PthAs on bacterial growth in planta. Leaves of sweet orange and lemon plants were infiltrated with bacterial suspensions (106 cells/mL) and the growth of wild‐type Xanthomonas citri (WT) and respective pthA‐deletion mutants (Δ) was monitored at 2 and 14 days after bacterial inoculation. The double (Δ1‐4 and Δ3‐4) and triple (Δ1‐3‐4) pthA‐deletion mutants grew significantly less well than the respective single mutants and the WT bacteria in all the citrus hosts tested, suggesting an additive effect of PthAs on bacterial growth in planta. Bacterial growth, expressed in colony‐forming units (CFU)/cm2 of leaf, is the mean of three biological replicates. The error bars denote standard deviations, whereas the asterisks above the bars indicate statistically significant differences between X. citri and mutant‐inoculated plants (P < 0.05).
PthA‐induced genes are differentially regulated in a host‐dependent manner
The evidence that PthAs 1 and 3 have additive effects on canker development elicited by PthA4 in ‘Pera’ sweet orange, but not ‘Tahiti’ lemon (Figs 1 and 2), suggested that PthAs 1 and 3 could activate additional canker S genes or help to enhance LOB1 expression in ‘Pera’ leaves. Thus, in addition to LOB1, we evaluated the expression levels of LOB2, LOB3 and DIOX, identified previously as putative PthA targets (Pereira et al., 2014). We found that LOB1 expression was significantly reduced in ‘Tahiti’ leaves infiltrated with each of the single pthA mutants, indicating that not only PthA4, but also PthAs 1 and 3, activate LOB1 transcription in this host (Fig. 4A). However, although PthA4 activates LOB1 in ‘Pera’ leaves, confirming previous results (Hu et al., 2014; Li et al., 2014; Pereira et al., 2014), PthA1 seems to repress LOB1 transcription in this host, as higher LOB1 expression levels were detected in ‘Pera’ leaves infiltrated with the Δ1 mutant relative to the wild‐type bacterium (Fig. 4A). No significant differences in LOB1 expression were observed in ‘Pera’ plants challenged with the Δ3 mutant relative to the wild‐type bacterium, suggesting that PthA3 induces LOB1 transcription only in ‘Tahiti’ plants (Fig. 4A).
Figure 4.
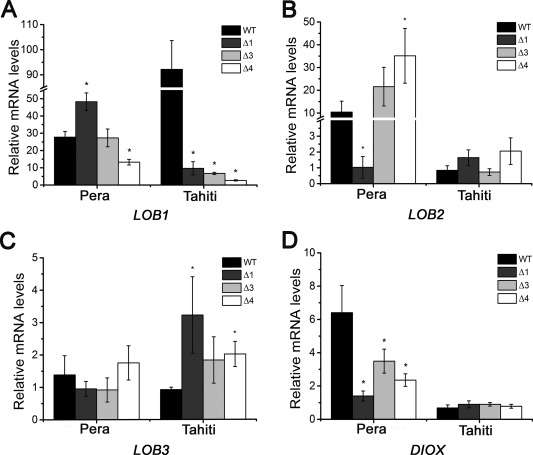
Differential PthA‐dependent induction of LOB (LATERAL ORGAN BOUNDARIES) and DIOX (citrus dioxygenase) genes in sweet orange and lemon plants. Expression levels of LOB1, LOB2, LOB3 and DIOX in ‘Pera’ and ‘Tahiti’ leaves infiltrated with the wild‐type (WT) Xanthomonas citri or the single pthA‐deletion mutants (Δ1, Δ3 and Δ4), 72 h post‐inoculation. (A) PthAs 1, 3 and 4 are required for LOB1 induction in lemon, whereas only PthA4 is important for LOB1 induction in ‘Pera’ sweet orange. However, PthA1 seems to repress LOB1 transcription in this host. (B, C) LOB2 and LOB3 are induced by X. citri in ‘Pera’ and ‘Tahiti’ plants; however, PthAs 1, 3 and 4 do not alter significantly LOB2 expression in ‘Tahiti’ or LOB3 expression in ‘Pera’ plants. LOB2 expression in ‘Pera’ sweet orange appears to require PthA1, but not PthA3 or PthA4. (D) The DIOX gene is also induced by X. citri WT and respective single pthA mutants in ‘Pera’ and ‘Tahiti’ leaves; however, deletion in pthA 1, 3 or 4 significantly reduces DIOX expression in ‘Pera’, but not in ‘Tahiti’, leaves. The error bars denote standard deviations, whereas the asterisks above the bars indicate statistically significant differences between X. citri and mutant‐inoculated plants (P < 0.05).
LOB2 (orange1.1g040761m.g) and LOB3 (orange1.1g036534m) were also induced by X. citri in ‘Pera’ sweet orange and ‘Tahiti’ lemon; however, PthAs 1, 3 and 4 do not seem to influence LOB2 expression in ‘Tahiti’ or LOB3 expression in ‘Pera’ plants (Fig. 4B,C). LOB2 expression in sweet orange appears to depend on PthA1, as significantly lower levels of LOB2 transcripts were detected in ‘Pera’ leaves in response to the Δ1 mutant relative to the wild‐type X. citri (Fig. 4B). On the other hand, and contrary to previous observations (Pereira et al., 2014), a deletion in pthA4 enhanced LOB2 expression in ‘Pera’ leaves. The slight increase in LOB3 expression in ‘Tahiti’ plants infiltrated with the single pthA mutants also suggests that PthAs 1, 3 and 4 repress LOB3 expression in this host (Fig. 4C).
The citrus DIOX gene (orange1.1g017949m), identified previously as a potential direct target of PthA4 and PthAw, is highly induced in citrus epicotyls expressing PthA4 (Hu et al., 2014; Pereira et al., 2014). Therefore, we tested whether PthA1 or PthA3 could also modulate DIOX transcription in ‘Pera’ relative to ‘Tahiti’ plants. We found that, although DIOX is up‐regulated by X. citri and by the single pthA mutants in ‘Pera’ and ‘Tahiti’ leaves, deletion in pthAs 1, 3 or 4 substantially reduces DIOX expression in ‘Pera’, but not ‘Tahiti’, leaves (Fig. 4D). These results confirm previous data showing that DIOX is induced in response to X. citri infection in ‘Pera’ leaves and that its expression is not altered by PthA2, nor entirely dependent on PthA4, in citrus epicotyls (Pereira et al., 2014). Taken together, these results indicate that LOB2 and DIOX might also function as canker S genes and that PthAs 1 and 3 contribute to their expression in ‘Pera’, but not ‘Tahiti’, plants.
LOB1 induction is not sufficient to elicit canker on citrus
We observed that, in spite of the high levels of LOB1 induced by the Δ1 and Δ3 mutants in ‘Pera’ compared with ‘Tahiti’ leaves, canker pustules developed normally in ‘Tahiti’, but were reduced in ‘Pera’ leaves, when challenged with the pthA1‐ and pthA3‐knockout mutants (Figs 1 and 4A). In addition, although LOB1 expression decreased by approximately 95% in ‘Tahiti’ compared with a 50% reduction in ‘Pera’ leaves infiltrated with the Δ4 mutant, canker lesions did not develop in any of these hosts (Figs 1 and 4A). These results suggest that there is either a threshold of LOB1 expression necessary to promote canker or that LOB1 induction alone is not sufficient for canker development in these hosts.
To investigate this idea, we measured the expression levels of LOB1 in ‘Pera’ sweet orange and ‘Mexican’ lime leaves infiltrated with X. aurantifolii ‘C’, in comparison with X. citri, as X. aurantifolii ‘C’ causes canker in ‘Mexican’ lime, but HR in ‘Pera’ (Fig. 5A) (Cernadas et al., 2008). We found, surprisingly, that LOB1 was weakly induced in the compatible interaction in ‘Mexican’ lime, and highly induced in both the compatible and incompatible interactions in ‘Pera’ (Fig. 5B). Therefore, LOB1 induction does not always correlate with canker development in citrus plants.
Figure 5.
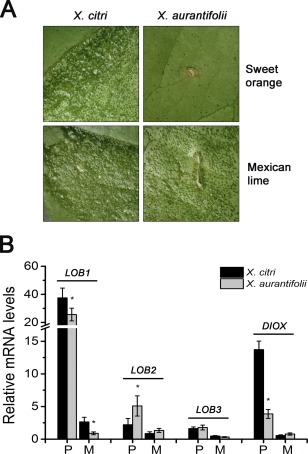
LOB1 (LATERAL ORGAN BOUNDARIES 1) induction does not always correlate with canker development. (A) Leaves of ‘Pera’ sweet orange and ‘Mexican’ lime were infiltrated with Xanthomonas citri and X. aurantifolii ‘C’, and canker symptoms were evaluated 14 days after bacterial infiltration (106 cells/mL). In contrast with a hypersensitive response (HR) elicited by X. aurantifolii ‘C’ in sweet orange compared with lemon, canker lesions developed in both ‘Pera’ and ‘Mexican’ lime inoculated with X. citri. (B) Expression levels of LOB1, LOB2, LOB3 and DIOX (citrus dioxygenase) genes in ‘Pera’ (P) relative to ‘Mexican’ lime (M) leaves in response to X. citri or X. aurantifolii ‘C’ infection. LOB1 is weakly induced in the compatible interactions in ‘Mexican’ lime and highly induced in both the compatible and incompatible interactions in ‘Pera’. The expression level of DIOX, but not of LOB1, LOB2 or LOB3, is significantly reduced in the incompatible interaction between ‘Pera’ and X. aurantifolii ‘C’. The error bars denote standard deviations, whereas the asterisks above the bars indicate statistically significant differences between X. citri‐ and X. aurantifolii‐inoculated plants (P < 0.05).
We also investigated the expression levels of LOB2, LOB3 and DIOX in ‘Pera’ and ‘Mexican’ lime leaves in response to X. citri or X. aurantifolii ‘C’ infection. Although LOB2 and LOB3 expression did not seem to correlate with either canker development or defence response in these hosts, the expression of DIOX was substantially reduced in the HR response elicited by X. aurantifolii ‘C’ in ‘Pera’ leaves (Fig. 5B). Together, these results suggest that LOB1 induction per se is not sufficient for canker formation and that DIOX might contribute to disease development.
CsDIOX is structurally related to 2OGDs
The protein encoded by the citrus DIOX gene belongs to the 2OGD family of enzymes that use iron and 2‐oxoglutarate as cofactors. Members of this family catalyse many different reactions, including hydroxylation, halogenation, demethylation, epimerization, among others (Farrow and Facchini, 2014; Martinez and Hausinger, 2015). In plants, 2OGDs play important roles in the biosynthesis and metabolism of numerous compounds, such as flavonoids, coumarins, glucosinolates, gibberellins, ethylene, auxin and salicylic acid (Farrow and Facchini, 2014).
The citrus DIOX protein (CsDIOX) is closely related to the Arabidopsis and tobacco feruloyl‐CoA 6′‐hydroxylase 1 (F6′H1), which catalyses the conversion of feruloyl‐CoA into 6′‐hydroxylferuloyl‐CoA during the synthesis of the coumarin scopoletin (Sun et al., 2014, 2015). To gain insights into the possible biochemical function and substrate specificity of CsDIOX, we generated a structural model of CsDIOX based on the Arabidopsis AtF6′H1 crystal structure (Sun et al., 2015). Our molecular model suggests that CsDIOX is structurally very similar to AtF6′H1 (Fig. 6A). In addition to having the typical protein fold and topology of the 2OGD family, all the amino acid residues responsible for the binding of the iron atom (H234, D236, H292), 2‐oxoglutarate (N217, Y219, R302, S304) and substrate feruloyl‐CoA (F150, S152, R213, N215, I237, F308) are structurally conserved in the CsDIOX model (Fig. 6B). These observations indicate that CsDIOX is the orthologue of AtF6′H1 (Kai et al., 2008; Sun et al., 2015) and possibly uses feruloyl‐CoA and p‐coumaroyl‐CoA as substrates.
Figure 6.
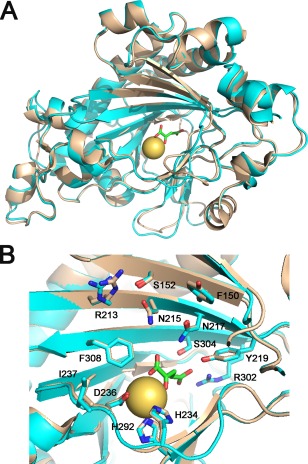
The citrus DIOX protein (CsDIOX) is structurally related to Arabidopsis feruloyl‐CoA 6′‐hydroxylase 1 (F6′H1). (A) Superposition of the CsDIOX structural model (cyan) with the Arabidopsis AtF6′H1 crystal structure (sand), depicting the iron atom (yellow) and 2‐oxoglutarate (green) (PDB code 4XAE; Sun et al., 2015), showing that CsDIOX displays the same folding and topology as AtF6′H1. (B) All of the amino acid residues responsible for the binding of the iron atom (H234, D236, H292), 2‐oxoglutarate (N217, Y219, R302, S304) and feruloyl CoA (F150, S152, R213, N215, I237, F308) are structurally conserved in the CsDIOX model.
Psoralen inhibits canker formation in ‘Pera’ sweet orange
CsDIOX is also 82% identical to the Ruta graveolens 2OGD, which converts feruloyl‐CoA into scopoletin and p‐coumaroyl‐CoA into 2,4‐dihydroxycinnamic acid (Vialart et al., 2012). As the results shown in Figs 4D and 5B suggest that DIOX could play a role in canker development, and psoralen, a furanocoumarin, inhibits the p‐coumaroyl‐ and feruloyl‐CoA hydroxylase activities of R. graveolens 2OGD at concentrations of 50–200 µm (Vialart et al., 2012), we tested the effect of 0.1 and 0.5 mm psoralen on X. citri growth and disease development. Notably, we found that psoralen inhibited canker formation and the growth of X. citri in a dose‐dependent manner in leaves of ‘Pera’ sweet orange, but not ‘Tahiti’ lemon (Fig. 7), where the expression of DIOX was not dependent on PthAs or significantly induced in response to X. citri infection (Fig. 4D). Taken together, these results suggest that citrus DIOX acts as a canker S gene in ‘Pera’ sweet orange.
Figure 7.
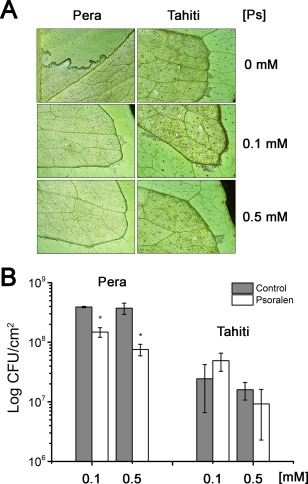
Psoralen inhibits canker formation in ‘Pera’ sweet orange. Leaves of ‘Pera’ sweet orange and ‘Tahiti’ lemon were infiltrated with a suspension of Xanthomonas citri (106 cells/mL) in the absence or presence of 0.1 or 0.5 mm psoralen (Ps). Canker symptoms (A) and bacterial counts (B) were evaluated 10 days after bacterial inoculation. Psoralen significantly inhibited canker development and bacterial growth in ‘Pera’ sweet orange, but not in ‘Tahiti’ lemon. Bacterial growth, expressed in colony‐forming units (CFU)/cm2 of leaf, is the mean of three biological replicates. The error bars denote standard deviations, whereas the asterisks above the bars indicate statistically significant differences between means (P < 0.05).
Promoters of LOB1, LOB2 and DIOX show polymorphisms at PthA sites
The differential PthA‐dependent expression of LOB1, LOB2 and DIOX genes observed between ‘Pera’ and ‘Tahiti’ plants suggests the existence of nucleotide polymorphisms at effector‐binding sites. Thus, we sequenced the promoter regions of these genes amplified from ‘Pera’ and ‘Tahiti’ plants and compared them with those of the published genomes of C. sinensis ‘Valencia’ and C. clementina ‘Clemenules’ (Wu et al., 2014; Xu et al., 2013).
In the LOB1 promoter, we found a single nucleotide change (C/T) and a single nucleotide deletion (G/–) flanking the characterized PthA4‐binding site (Hu et al., 2014; Pereira et al., 2014) among the ‘Tahiti’, ‘Valencia’, ‘Pera’ and ‘Clemenules’ promoters (Fig. 8A). We also found an 11‐nucleotide deletion 25 bp downstream of the PthA4 site in the ‘Tahiti’ relative to the ‘Pera’, ‘Valencia’ or ‘Clemenules’ promoters. In addition to the PthA4 site, we found putative PthA1 and PthA3 sites located 667 and 498 bp upstream of the ATG, respectively, both carrying single nucleotide polymorphisms (SNPs) between the ‘Pera’ (A/C) and ‘Tahiti’ (A/C) promoters (Fig. 8A).
Figure 8.
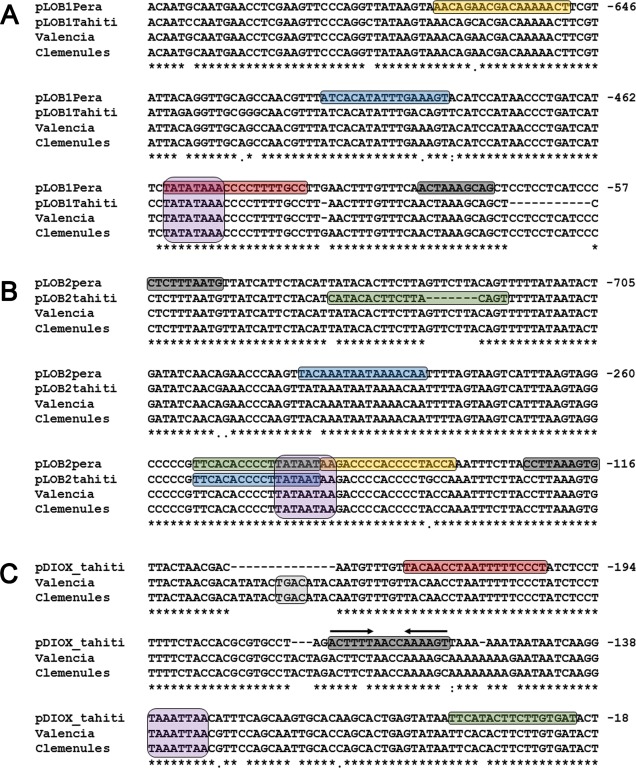
Promoters of LOB1 (LATERAL ORGAN BOUNDARIES 1), LOB2 and DIOX (citrus dioxygenase) genes show nucleotide polymorphisms at PthA sites. The promoter regions of the citrus LOB1 (A), LOB2 (B) and DIOX (C) genes were amplified from ‘Pera’ and ‘Tahiti’ plants and compared with those of the Citrus sinensis ‘Valencia’ and C. clementine ‘Clemenules’ cultivars (Wu et al., 2014; Xu et al., 2013). The predicted PthA1‐, PthA2‐, PthA3‐ and PthA4‐binding sites are boxed in yellow, green, blue and red, respectively. The Dof‐ and WRKY‐binding sites are coloured in grey and light grey, respectively, whereas the TATA box elements are in purple. Arrows in (C) represent a Dof palindromic sequence. Most of the single nucleotide polymorphisms (SNPs) found in ‘Pera’ relative to ‘Tahiti’ plants are also present in ‘Valencia’ and ‘Clemenules’ cultivars.
The LOB2 promoter also seems to cluster multiple PthA sites. In addition to the PthA2 site identified previously (Pereira et al., 2014), which overlaps with the predicted TATA box element, we found additional PthA1, PthA2 and PthA3 sites which also present nucleotide deletions or SNPs between the ‘Pera’ and ‘Tahiti’ sequences (Fig. 8B).
Despite many efforts, the DIOX promoter could not be amplified from several ‘Pera’ individuals, even though we tested six distinct pairs of oligos derived from the ‘Valencia’ genome (Table S1, see Supporting Information) in multiple polymerase chain reaction (PCRs). This suggests that this genomic region is highly polymorphic. Indeed, when we compared the sequence of the DIOX promoter obtained from ‘Tahiti’ plants with those of ‘Valencia’ and ‘Clemenules’, we found nucleotide insertions ranging from 150 to 1230 bp located at approximately 270 bp upstream of the ATG (not shown). Thus, the −260‐bp sequence of the ‘Tahiti’ promoter was aligned with the corresponding sequences of the ‘Valencia’ and ‘Clemenules’ promoters (Fig. 8C). In addition to the PthA4 site described previously (Pereira et al., 2014), we found a PthA2 site that also shows an SNP between the ‘Tahiti’ and ‘Valencia’ or ‘Clemenules’ promoters (Fig. 8C). Moreover, as observed in the LOB1 promoter, there is a 14‐bp deletion flanking the PthA4 site in the ‘Tahiti’ relative to the ‘Valencia’ or ‘Clemenules’ promoters (Fig. 8C). According to the PlantPAN 2.0 algorithm (Chang et al., 2008), this deletion comprise a ‘TGAC’ core‐containing W box which is bound by WRKY factors that control pathogenesis‐related and gibberellin response genes (Eulgem et al., 1999; Zhang et al., 2004).
In addition to the predicted W box found in the DIOX promoter, a PlantPAN 2.0 search identified several putative transcription factor‐binding sites in the LOB1, LOB2 and DIOX promoters. One particular factor, Dof, attracted our attention, because Dof sites were found near PthA sites in all three promoters (Fig. 8). Most notably, in the DIOX promoter, two inverted Dof sites form a palindrome located 28 bp downstream of the PthA4 site (Fig. 8C). We also observed single nucleotide changes or deletions near or at the Dof sites between the sweet orange and lemon sequences (Fig. 8). Together, these results indicate that the differential PthA‐dependent expression of LOB1, LOB2 and DIOX observed between ‘Pera’ and ‘Tahiti’ plants might be influenced by the polymorphisms found at the PthA sites, and that WRKY and Dof factors might play an important role in the transcriptional regulation of these genes.
Discussion
Although many Xanthomonas pathogens, including X. citri and X. oryzae, carry multiple TAL effector genes, the precise role or contribution of each of these effectors to pathogenicity is poorly understood, as only a few have been shown to act as major virulence factors (Al‐Saadi et al., 2007; Wilkins et al., 2015; Yang and White, 2004). In the case of X. citri strain 306, which carries four PthA effectors, only PthA4 is known to play a fundamental role in canker elicitation, whereas none seem to act as avirulence factors on citrus (Al‐Saadi et al., 2007; Hu et al., 2014; da Silva et al., 2002; Yan and Wang, 2012).
In this study, we have shown that, although PthA4 is indispensable for canker formation in all citrus varieties tested, corroborating literature data (Al‐Saadi et al., 2007; Hu et al., 2014; Soprano et al., 2013; Yan and Wang, 2012), PthAs 1 and 3 also contribute to disease symptom development and bacterial growth in some citrus hosts, including ‘Pera’ sweet orange. These results indicate that TAL effectors of X. citri have a host‐dependent additive or complementary role in pathogenicity, and suggest that PthAs 1 and 3 potentiate the role played by PthA4 in canker elicitation. This idea is consistent with the fact that PthAs 1 and 3 are not required for LOB1 induction in ‘Pera’ plants, but for the activation of other targets, including LOB2 and DIOX. Importantly, we show that LOB1 induction by X. citri does not always correlate with canker formation. The expression of LOB1 is, for instance, drastically reduced in ‘Tahiti’ leaves infiltrated with the pthA‐deletion mutants (Fig. 4A), although canker lesions develop normally in this host relative to ‘Pera’ sweet orange (Figs 1 and 2). Conversely, LOB1 is strongly up‐regulated during the incompatible interaction between X. aurantifolii ‘C’ and ‘Pera’ plants (Fig. 5). These data indicate that LOB1 induction per se is not sufficient for canker elicitation, which is in agreement with the observation that the transient overexpression of LOB1 in transgenic citrus does not result in the formation of canker pustules (Hu et al., 2014). Therefore, although LOB1 appears to directly up‐regulate many cell wall‐remodelling genes which are also induced by PthA4 during canker development (Hu et al., 2014), it is unlikely that LOB1 activation alone would trigger the whole process of canker formation.
In line with this idea, we present evidence suggesting that citrus DIOX, identified as a PthA4 target in two independent studies (Hu et al., 2014; Pereira et al., 2014), might also play an important role in canker development. The expression of DIOX is dependent on PthAs 1, 3 and 4 in ‘Pera’ sweet orange, but not in ‘Tahiti’ lemon (Fig. 4D). In addition, psoralen, an inhibitor of 2OGD (Vialart et al., 2012), significantly attenuates canker formation and bacterial growth in ‘Pera’, but not ‘Tahiti’, plants (Fig. 7). Although the biological function and substrate specificity of CsDIOX have not yet been demonstrated, our molecular modelling studies strongly suggest that CsDIOX is the orthologue of the Arabidopsis and R. graveolens F6′H1 enzymes (Fig. 6), and can thus catalyse the ortho‐hydroxylation of feruloyl‐CoA and/or p‐coumaroyl‐CoA (Kai et al., 2008; Sun et al., 2015; Vialart et al., 2012). The question that arises is therefore how the CsDIOX activity could lead or contribute to citrus canker development in ‘Pera’ sweet orange.
One possible mechanism might involve the action of cinnamoyl‐CoA reductases (CCRs), which convert cinnamoyl‐CoA esters, including feruloyl‐CoA, p‐coumaroyl‐CoA and caffeoyl‐CoA, into their corresponding cinnamyl aldehydes during the synthesis of lignin monomers (Xue et al., 2015; Zhou et al., 2010). It has been shown that the Arabidopsis CCR1 enzyme mediates the cell proliferation exit for leaf development and that the ccr1 mutant exhibits increased cell proliferation and high levels of ferulic acid (Xue et al., 2015). Because CCR1 preferentially uses feruloyl‐CoA as substrate, it is possible that 6‐hydroxylferuloyl‐CoA, the product of the CsDIOX reaction, inhibits the activity of CCR1, leading to a cell proliferation response similar to that of the loss of CCR1 (Xue et al., 2015; Zhou et al., 2010). Moreover, the role of CCR1 proteins in lignin biosynthesis and cell division is also linked to the activities of caffeic acid O‐methyltransferases (COMTs) and caffeoyl‐CoA 3‐O‐methyltransferases (CCOAOMTs), which provide the substrates for CCR1 (Xue et al., 2015; Zhou et al., 2010). Inhibition of COMT and CCOAOMT in Arabidopsis results in low levels of ferulic acid and decreased cell division (Xue et al., 2015). As ferulic acid itself induces cell proliferation in Arabidopsis, it would be interesting to investigate whether 6‐hydroxylferulic acid can also stimulate cell division during canker formation.
It is also notable that many COMT, CCOAOMT and CCR genes are preferentially up‐regulated by X. aurantifolii ‘C’ during its incompatible interaction in ‘Pera’ sweet orange (Cernadas et al., 2008). We initially interpreted this pattern of COMT, CCOAOMT and CCR gene induction as part of the X. aurantifolii‐elicited HR, in which increased lignin biosynthesis reinforces the plant cell wall to restrict pathogen attack (Cernadas et al., 2008). However, in the light of the data reported by Xue et al. (2015), it is possible that the citrus COMT, CCOAOMT and CCR pathway also helps to restrict cell division during canker formation. The fact that the citrus DIOX gene is induced in ‘Pera’ leaves at much lower levels by X. aurantifolii ‘C’ relative to X. citri (Fig. 5B) is consistent with the idea that DIOX plays an opposing role in the COMT, CCOAOMT and CCR pathway. Moreover, the observation that PthA4 up‐regulates a second F6′H1 gene (XP_006488907.1) and that EBEs for PthAs are found in a number of citrus DIOX‐related 2OGD genes (Pereira et al., 2014) also suggests that this lignin biosynthetic pathway is differentially modulated by X. citri, leading to cell hyperplasia.
The differential PthA‐dependent expression of LOB1, LOB2 and DIOX observed between ‘Pera’ sweet orange and ‘Tahiti’ lemon plants is not only associated with the presence or absence of specific PthA sites in the promoters of these genes, but also with the occurrence of SNPs or nucleotide deletions found adjacent to or within such PthA sites (Fig. 8). A single nucleotide deletion in the X. oryzae PthXo2 EBE in the rice bacterial blight S gene Xa25 (OsSWEET13) is, for instance, sufficient to abrogate effector‐associated gene activation (Zhou et al., 2015). In another rice bacterial blight S gene, OsSWEET14, an 18‐bp deletion within the binding sites targeted by several TAL effectors also confers resistance against many X. oryzae strains of various geographical origins (Hutin et al., 2015). Therefore, the presence of nucleotide polymorphisms at PthA sites is strong evidence of a host adaptation to evade effector target recognition.
Near the PthA sites, we also found several putative Dof sites in the LOB1, LOB2 and DIOX promoters. Dof factors control the transcription of many plant genes. In Arabidopsis, Dof5.8 represses the auxin response, leading to impaired vein formation in leaves (Konishi and Yanagisawa, 2015), whereas DAG1 negatively regulates the gibberellin biosynthetic gene AtGA3ox1, which encodes a CsDIOX‐related 2OGD (Gabriele et al., 2010). In maize, Dof factors interact with HMGB (high‐mobility group B) proteins, which facilitate Dof binding to naked DNA or to nucleosomes (Cavalar et al., 2003; Grasser et al., 2007; Krohn et al., 2002; Yanagisawa, 1997).
The interaction between Dof and HMGBs is of particular note, as all PthAs from strain 306 interact with the citrus HMGB1 protein (de Souza et al., 2012). Mammalian HMGB1 interacts with the TATA‐binding protein (TBP) to repress transcription, whereas transcription factor TFIIA binds TBP to disrupt the HMGB/TBP complex and activate transcription (Das and Scovell, 2001; Dasgupta and Scovell, 2003; Sutrias‐Grau et al., 1999). Considering that Dof factors act as transcription repressors and that PthAs bind at or close to TATA box elements of citrus promoters, it is possible that, similar to TFIIA, PthAs could displace Dof/HMGB or alter the recruitment of factors at TBP sites to promote transcription initiation.
Experimental Procedures
Bacterial strains and growth conditions
Escherichia coli DH5α cells were grown in Luria–Bertani (LB) medium at 37 °C for 16 h, whereas X. citri strain 306 (da Silva et al., 2002) and X. aurantifolii ‘C’ strain ICMP 8435 (Cernadas et al., 2008) were grown in LB medium without NaCl (LBON) at 28 °C for 48 h. When required, ampicillin (100 µg/mL) and/or kanamycin (50 µg/mL) were added to the growth medium.
Construction of pthA‐deletion mutants
The single (Δ1, Δ3), double (Δ1‐3, Δ1‐4, Δ3‐4) and triple (Δ1‐3‐4) pthA‐knockout mutants of X. citri were obtained by homologous recombination following the same procedure as used to obtain the pthA4‐deletion mutant (Δ4), described previously (Soprano et al., 2013). To generate the Δ1 and Δ3 mutants, DNA fragments of ∼1.0 kb, flanking the coding regions of pthA1 and pthA3, were amplified using the pairs of oligos PthA1F1/PthANR1 and PthANF2/PthA1R2, and PthA3F1/PthANR1 and PthANF2/PthA3R2, respectively (Table S1 and Fig. 1A). The PCR fragments corresponding to the upstream and downstream regions of each pthA gene were ligated through the NdeI sites and cloned into the HindIII and EcoRI sites of the suicide vector pNPTS138 to create an in‐frame deletion (Andrade et al., 2014). The constructs were verified by DNA sequencing and used to transform X. citri cells by electroporation. Bacterial colonies were selected on LBON plates supplemented with ampicillin and kanamycin, or with ampicillin and 5% sucrose. Kanamycin‐sensitive colonies that grew in the presence of sucrose were selected and PCR tested for the absence of pthA genes after two recombination events (Fig. S1B, see Supporting Information). Despite many efforts, we were unable to select a pthA2‐deletion mutant. To confirm the pthA1 and pthA3 knockouts, the DNA fragments encompassing the deleted loci were amplified with oligos PthA1F1/PthA1R2 and PthA3F1/PthA3R2, respectively, and sequenced with oligo seqPthANF, which covers the in‐frame deletion regions (Table S1 and Fig. S1A, C).
The double (Δ1‐3, Δ1‐4, Δ3‐4) and triple (Δ1‐3‐4) pthA‐deletion mutants were generated using the single mutants in successive rounds of deletion. The double and triple pthA mutants were also confirmed by PCR analysis and DNA sequencing (Fig. S1B).
Plant growth and bacterial inoculations
The sweet orange (‘Pera’, ‘Valencia’, ‘Hamlin’, ‘Sorocaba’ and ‘Natal’) and lemon (‘Tahiti’ and ‘Mexican’ lime) varieties were obtained from certified nurseries as ‘pathogen free’ and maintained in glasshouse conditions. Leaves were infiltrated with water suspensions of 105 or 106 cells/mL of X. citri, X. aurantifolii ‘C’ or the pthA‐deletion mutants, previously grown on LBON plates for 48 h at 28 ºC. The wild‐type bacteria and corresponding pthA‐deletion mutants were infiltrated in nine distinct leaves from three independent plants of the same citrus variety. The inoculated plants were monitored daily for the appearance of canker symptoms.
Bacterial growth analysis
The growth of X. citri and pthA‐deletion mutants in leaf tissues was evaluated as described previously (Cernadas et al., 2008). Discs of leaf sectors infiltrated with approximately 106 bacterial cells were removed at 2 and 14 days after bacterial inoculation and ground in 1 mL of sterile water. Serial dilutions of the bacterial suspensions were plated on LBON supplemented with ampicillin, and bacterial colonies were counted from three independent leaf extractions. The experiment was repeated three times and statistically significant differences between treatments (X. citri vs. mutant‐inoculated plants) were calculated using Student's t‐test with P < 0.05.
Plant RNA extraction and reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) analysis
Total RNA extraction and RT‐qPCR analyses were conducted as described previously (Pereira et al., 2014). The oligonucleotides used as primers were designed using Applied Biosystems (Foster City, CA, USA) Primer Express 2.0 software (Table S1). PCR amplifications were carried out using the 7500 system ‘Universal’ cycle condition in an ABI Prism 7300 instrument (Applied Biosystems), and the gene encoding citrus actin was used as internal control for normalization (Mafra et al., 2012). Total RNAs from three different leaves were used in the PCRs as independent biological replicates, and three technical replicates for each biological sample were considered for statistical two‐tailed Student's t‐test to compare the changes in gene expression between treatments. The relative gene expression levels among samples were determined using 7000 System SDS software (Applied Biosystems) with default parameters.
Psoralen treatment
Leaves of ‘Pera’ sweet orange and ‘Tahiti’ lemon were infiltrated with a water suspension of X. citri (106 cells/mL) in the absence or presence of psoralen. Psoralen at 50 mm was dissolved in 100% ethanol and added to the bacterial cell suspensions at 0.1 and 0.5 mm final concentrations. Ethanol was also added to the bacterial cell suspensions at 0.2% or 1.0% final concentrations to serve as controls for psoralen treatments. Psoralen or ethanol at the concentrations used did not affect the growth of X. citri in culture medium (not shown). To maintain the active pools of the inhibitor, psoralen or ethanol was infiltrated into the inoculated leaf sectors on the third and sixth days of bacterial inoculation. The experiment was performed using nine infiltrated leaves from three plants of each citrus cultivar. Canker symptoms and bacterial counts were evaluated at 10 days after bacterial infiltration. Statistically significant differences between treatments were calculated using Student's t‐test with P < 0.05.
Cloning and sequencing of promoter regions of citrus genes
Genomic DNA of ‘Pera’ sweet orange and ‘Tahiti’ lemon was extracted from leaves using the cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle, 1987). The quality and quantity of DNA were verified by gel electrophoresis and UV spectroscopy. The promoter regions of LOB1, LOB2 and DIOX were amplified by PCR using 1.0 µg of genomic DNA, 125 nm of each primer and Platinum Taq, following the recommendations of the manufacturer (Thermo Fisher Scientific Inc., Waltham, MA). The oligonucleotides used as primers for LOB1, LOB2 and DIOX (Table S1) were designed based on the DNA sequences of the citrus genomes (Wu et al., 2014; Xu et al., 2013). After amplification (1 × 94 ºC for 4 min, followed by 35 × 94 ºC for 30 s, 55 ºC for 30 s and 72 ºC for 70 s), the PCR fragments were cloned into the pGEM T‐Easy vector (Promega, Madison, WI, USA) and sequenced. The promoter sequences were aligned using Clustal Omega software at http://www.ebi.ac.uk/Tools/msa/clustalo/.
Molecular modelling studies
The three‐dimensional structural model of CsDIOX was generated with the SWISS‐MODEL server at http://swissmodel.expasy.org/, applying default parameters and using the Arabidopsis thaliana AtF6′H1 crystal structure (PDB code 4XAE; Sun et al., 2015) as the search model. Structural alignments of the CsDIOX model with AtF6′H1 were performed with PyMOL (DeLano, 2002).
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Construction of pthA‐deletion mutants. (A) Strategy used for the deletion of the pthA genes in Xanthomonas citri strain 306. Schematic view of pthA1, pthA3 and pthA4 loci depicting the oligonucleotides (arrows) used to generate the 1.0‐kb fragments flanking the pthA coding regions (blue). The oligonucleotides PthA1F1, PthA1R2, PthA3F1, PthA3R2, PthA4F1 and PthA4R2 are specific for each pthA locus, whereas PthANR1 and PthANF2 are common to all pthAs. The bars below represent the approximate length of the DNA fragments. (B) Confirmation of the pthA deletions by polymerase chain reaction (PCR). DNAs from the single (Δ1, Δ3 and Δ4), double (Δ1‐3, Δ1‐4 and Δ3‐4) and triple (Δ1‐3‐4) mutants were amplified using the indicated oligonucleotides shown in (A) and resolved on agarose gel. The approximately 2.0‐kb PCR bands corresponding to the 1.0‐kb fragments flanking the pthA genes are indicative of the specific pthA deletions in each mutant background. (C) Examples of sequencing chromatograms of Δ1, Δ3 and Δ4 mutants showing the respective in‐frame pthA deletions. The amino acid sequences encoded by the mutated pthA loci are indicated. The 2.0‐kb fragments obtained with the specific PthAF and PthAR oligos were sequenced using oligo seqPthANF.
Table S1 Oligonucleotides used for the gene expression analyses, promoter cloning and sequencing, and pthA‐deletion mutant construction and verification.
Acknowledgements
This work was supported by the São Paulo Research Foundation (FAPESP Grants 2011/20468‐1 and 2011/19988‐0) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq Grant 300937/2012‐1).
References
- Al‐Saadi, A. , Reddy, J. , Duan, Y. , Brunings, A. , Yuan, Q. and Gabriel, D. (2007) All five host‐range variants of Xanthomonas citri carry one pthA homolog with 17.5 repeats that determines pathogenicity on citrus, but none determine host‐range variation. Mol. Plant–Microbe Interact. 20, 934–943. [DOI] [PubMed] [Google Scholar]
- Andrade, M.O. , Farah, C.S. and Wang, N. (2014) The post‐transcriptional regulator rsmA/csrA activates T3SS by stabilizing the 59 UTR of hrpG, the master regulator of hrp/hrc genes, in Xanthomonas . PLoS Pathog. 10, e1003945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony, G. , Zhou, J. , Huang, S. , Li, T. , Liu, B. , White, F. and Yang, B. (2010) Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os‐11N3. Plant Cell, 22, 3864–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch, J. and Bonas, U. (2010) Xanthomonas AvrBs3 family‐type III effectors: discovery and function. Annu. Rev. Phytopathol. 48, 419–436. [DOI] [PubMed] [Google Scholar]
- Boch, J. , Scholze, H. , Schornack, S. , Landgraf, A. , Hahn, S. , Kay, S. , Lahaye, T. , Nickstadt, A. and Bonas, U. (2009) Breaking the code of DNA binding specificity of TAL‐type III effectors. Science, 326, 1509–1512. [DOI] [PubMed] [Google Scholar]
- Brunings, A.M. and Gabriel, D.W. (2003) Xanthomonas citri: breaking the surface. Mol. Plant Pathol. 4, 141–157. [DOI] [PubMed] [Google Scholar]
- Cavalar, M. , Möller, C. , Offermann, S. , Krohn, N.M. , Grasser, K.D. and Peterhänsel, C. (2003) The interaction of DOF transcription factors with nucleosomes depends on the positioning of the binding site and is facilitated by maize HMGB5. Biochemistry, 42, 2149–2157. [DOI] [PubMed] [Google Scholar]
- Cernadas, R.A. , Camillo, L.R. and Benedetti, C.E. (2008) Transcriptional analysis of the sweet orange interaction with the citrus canker pathogens Xanthomonas axonopodis pv. citri and Xanthomonas axonopodis pv. aurantifolii . Mol. Plant Pathol. 9, 609–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, W.C. , Lee, T.Y. , Huang, H.D. , Huang, H.Y. and Pan, R.L. (2008) PlantPAN: Plant Promoter Analysis Navigator, for identifying combinatorial cis‐regulatory elements with distance constraint in plant gene group. BMC Genomics, 9, 561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiesa, M.A. , Siciliano, M.F. , Ornella, L. , Roeschlin, R.A. , Favaro, M.A. , Delgado, N.P. , Sendín, L.N. , Orce, I.G. , Ploper, L.D. , Vojnov, A.A. , Vacas, J.G. , Filippone, M.P. , Castagnaro, A.P. and Marano, M.R. (2013) Characterization of a variant of Xanthomonas citri subsp. citri that triggers a host‐specific defense response. Phytopathology, 103, 555–564. [DOI] [PubMed] [Google Scholar]
- Das, D. and Scovell, W.M. (2001) The binding interaction of HMG‐1 with the TATA‐binding protein/TATA complex. J. Biol. Chem. 276, 32 597–32 605. [DOI] [PubMed] [Google Scholar]
- Dasgupta, A. and Scovell, W.M. (2003) TFIIA abrogates the effects of inhibition by HMGB1 but not E1A during the early stages of assembly of the transcriptional preinitiation complex. Biochim. Biophys. Acta, 1627, 101–110. [DOI] [PubMed] [Google Scholar]
- DeLano, W.L. (2002) The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific. [Google Scholar]
- Deng, D. , Yan, C. , Pan, X. , Mahfouz, M. , Wang, J. , Zhu, J.K. , Shi, Y. and Yan, N. (2012) Structural basis for sequence‐specific recognition of DNA by TAL effectors. Science, 335, 720–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues, M.N. , Souza, T.A. , Cernadas, R.A. , de Oliveira, M.L.P. , Docena, C. , Farah, C.S. and Benedetti, C.E. (2010) The Xanthomonas citri effector protein PthA interacts with citrus proteins involved in nuclear transport, protein folding and ubiquitination associated with DNA repair. Mol. Plant Pathol. 11, 663–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, J.J. and Doyle, J.L. (1987) A rapid isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15. [Google Scholar]
- Duan, Y. , Castañeda, A. , Zhao, G. , Erdos, G. and Gabriel, D. (1999) Expression of a single, host‐specific, bacterial pathogenicity gene in plant cells elicits division, enlargement, and cell death. Mol. Plant–Microbe Interact. 12, 556–560. [Google Scholar]
- Eulgem, T. , Rushton, P.J. , Schmelzer, E. , Hahlbrock, K. and Somssich, I.E. (1999) Early events in plant defence signaling: rapid gene activation by WRKY transcription factors. EMBO J. 18, 4689–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow, S.C. and Facchini, P.J. (2014) Functional diversity of 2‐oxoglutarate/Fe(II)‐dependent dioxygenases in plant metabolism. Front. Plant Sci. 5, 524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriele, S. , Rizza, A. , Martone, J. , Circelli, P. , Costantino, P. and Vittorioso, P. (2010) The DOF protein DAG1 mediates PIL5 activity on seed germination by negatively regulating the GA biosynthetic gene AtGA3ox1 . Plant J. 61, 312–323. [DOI] [PubMed] [Google Scholar]
- Graham, J.H. , Gottwald, T.R. , Cubero, J. and Achor, D.S. (2004) Xanthomonas axonopodis pv. citri: factors affecting successful eradication of citrus canker. Mol. Plant Pathol. 5, 1–15. [DOI] [PubMed] [Google Scholar]
- Grasser, M. , Christensen, J.M. , Peterhänsel, C. and Grasser, K.D. (2007) Basic and acidic regions flanking the HMG‐Box domain of maize HMGB1 and HMGB5 modulate the stimulatory effect on the DNA binding of transcription factor Dof2. Biochemistry, 46, 6375–6382. [DOI] [PubMed] [Google Scholar]
- Hu, Y. , Zhang, J. , Jia, H. , Sosso, D. , Li, T. , Frommer, W.B. , Yang, B. , White, F.F. , Wang, N. and Jones, J.B. (2014) Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. USA, 111, 521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutin, M. , Sabot, F. , Ghesquière, A. , Koebnik, R. and Szurek, B. (2015) A knowledge‐based molecular screen uncovers a broad spectrum OsSWEET14 resistance allele to bacterial blight from wild rice. Plant J. 84, 694–703. doi: 10.1111/tpj.13042. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Kai, K. , Mizutani, M. , Kawamura, N. , Yamamoto, R. , Tamai, M. , Yamaguchi, H. , Sakata, K. and Shimizu, B. (2008) Scopoletin is biosynthesized via orthohydroxylation of feruloyl CoA by a 2‐oxoglutarate‐dependent dioxygenase in Arabidopsis thaliana . Plant J. 55, 989–999. [DOI] [PubMed] [Google Scholar]
- Kay, S. , Hahn, S. , Marois, E. , Hause, G. and Bonas, U. (2007) A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science, 318, 648–651. [DOI] [PubMed] [Google Scholar]
- Konishi, M. and Yanagisawa, S. (2015) Transcriptional repression caused by Dof5.8 is involved in proper vein network formation in Arabidopsis thaliana leaves. J. Plant Res. 128, 643–652. [DOI] [PubMed] [Google Scholar]
- Krohn, N.M. , Yanagisawa, S. and Grasser, K.D. (2002) Specificity of the stimulatory interaction between chromosomal HMGB proteins and the transcription factor Dof2 and its negative regulation by protein kinase CK2‐mediated phosphorylation. J. Biol. Chem. 277, 32 438–32 444. [DOI] [PubMed] [Google Scholar]
- Li, Z. , Zou, L. , Ye, G. , Xiong, L. , Ji, Z. , Zakria, M. , Hong, N. , Wang, G. and Chen, G. (2014) A potential disease susceptibility gene CsLOB of citrus is targeted by a major virulence effector PthA of Xanthomonas citri subsp. citri . Mol. Plant. 7, 912–915. [DOI] [PubMed] [Google Scholar]
- Mafra, V. , Kubo, K.S. , Alves‐Ferreira, M. , Ribeiro‐Alves, M. , Stuart, R.M. , Boava, L.P. , Rodrigues, C.M. and Machado, M.A. (2012) Reference genes for accurate transcript normalization in citrus genotypes under different experimental conditions. PLoS One, 7, e31263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak, A.N.S. , Bradley, P. , Cernadas, R.A. , Bogdanove, A.J. and Stoddard, B.L. (2012) The crystal structure of TAL effector PthXo1 bound to its DNA target. Science, 335, 716–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, S. and Hausinger, R.P. (2015) Catalytic mechanisms of Fe(II)‐ and 2‐oxoglutarate‐dependent oxygenases. J. Biol. Chem. 290, 20 702–20 711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou, M. and Bogdanove, A. (2009) A simple cipher governs DNA recognition by TAL effectors. Science, 326, 1501. [DOI] [PubMed] [Google Scholar]
- Murakami, M.T. , Sforça, M.L. , Neves, J.L. , Paiva, J.H. , Domingues, M.N. , Pereira, A.L.A. , Zeri, A.C.M. and Benedetti, C.E. (2010) The repeat domain of the type III effector protein PthA shows a TPR‐like structure and undergoes conformational changes upon DNA interaction. Proteins, 78, 3386–3395. [DOI] [PubMed] [Google Scholar]
- Pereira, A.L. , Carazzolle, M.F. , Abe, V.Y. , de Oliveira, M.L. , Domingues, M.N. , Silva, J.C. , Cernadas, R.A. and Benedetti, C.E. (2014) Identification of putative TAL effector targets of the citrus canker pathogens shows functional convergence underlying disease development and defense response. BMC Genomics, 15, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römer, P. , Hahn, S. , Jordan, T. , Strauss, T. , Bonas, U. and Lahaye, T. (2007) Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science, 318, 645–648. [DOI] [PubMed] [Google Scholar]
- Shiotani, H. , Fujikawa, T. , Ishihara, H. , Tsuyumu, S. and Ozaki, K. (2007) A pthA homolog from Xanthomonas axonopodis pv. citri responsible for host‐specific suppression of virulence. J. Bacteriol. 189, 3271–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shubert, T.S. , Rizvi, S.A. , Sun, X. , Gottwald, T.R. , Graham, J.H. and Dixon, W.N. (2003) Meeting the challenge of eradicating citrus canker in Florida again. Plant Dis. 85, 340–356. [DOI] [PubMed] [Google Scholar]
- da Silva, A. , Ferro, J. , Reinach, F.C. , Farah, C.S. , Furlan, L.R. , Quaggio, R.B. , Monteiro‐Vitorello, C.B. , Van Sluys, M.A. , Almeida, N.F. , Alves, L.M. , do Amaral, A.M. , Bertolini, M.C. , Camargo, L.E. , Camarotte, G. , Cannavan, F. , Cardozo, J. , Chambergo, F. , Ciapina, L.P. , Cicarelli, R.M. , Coutinho, L.L. , Cursino‐Santos, J.R. , El‐Dorry, H. , Faria, J.B. , Ferreira, A.J. , Ferreira, R.C. , Ferro, M.I. , Formighieri, E.F. , Franco, M.C. , Greggio, C.C. , Gruber, A. , Katsuyama, A.M. , Kishi, L.T. , Leite, R.P. , Lemos, E.G. , Lemos, M.V. , Locali, E.C. , Machado, M.A. , Madeira, A.M. , Martinez‐Rossi, N.M. , Martins, E.C. , Meidanis, J. , Menck, C.F. , Miyaki, C.Y. , Moon, D.H. , Moreira, L.M. , Novo, M.T. , Okura, V.K. , Oliveira, M.C. , Oliveira, V.R. , Pereira, H.A. , Rossi, A. , Sena, J.A. , Silva, C. , de Souza, R.F. , Spinola, L.A. , Takita, M.A. , Tamura, R.E. , Teixeira, E.C. , Tezza, R.I. , Trindade dos Santos, M. , Truffi, D. , Tsai, S.M. , White, F.F. , Setubal, J.C. and Kitajima, J.P. (2002) Comparison of the genomes of two Xanthomonas pathogens differing in host specificities. Nature, 417, 459–463. [DOI] [PubMed] [Google Scholar]
- Soprano, A.S. , Abe, V.Y. , Smetana, J.H. and Benedetti, C.E. (2013) Citrus MAF1, a repressor of RNA Pol III, binds the Xanthomonas citri canker elicitor PthA4 and suppresses citrus canker development. Plant Physiol. 163, 232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza, T.A. , Soprano, A.S. , de Lira N.P.V., Quaresma, A.J.C. , Pauletti, B.A. , Leme, A.F.P. and Benedetti, C.E. (2012) The TAL effector PthA4 interacts with nuclear factors involved in RNA dependent processes including a HMG protein that selectively binds poly(U) RNA. PLoS One, 7, e32305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H. , Wang, L. , Zhang, B. , Ma, J. , Hettenhausen, C. , Cao, G. , Sun, G. , Wu, J. and Wu, J. (2014) Scopoletin is a phytoalexin against Alternaria alternata in wild tobacco dependent on jasmonate signalling. J. Exp. Bot. 65, 4305–4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X. , Zhou, D. , Kandavelu, P. , Zhang, H. , Yuan, Q. , Wang, B.C. , Rose, J. and Yan, Y. (2015) Structural insights into substrate specificity of feruloyl‐CoA 6′‐hydroxylase from Arabidopsis thaliana . Sci. Rep. 5, 10 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutrias‐Grau, M. , Bianchi, M.E. and Bernue's, J. (1999) High mobility group protein 1 interacts specifically with the core domain of human TATA box‐binding protein and interferes with transcription factor IIB within the pre‐initiation complex. J. Biol. Chem. 274, 1628–1634. [DOI] [PubMed] [Google Scholar]
- Swarup, S. , Yang, Y. , Kingsley, M.T. and Gabriel, D.W. (1992) An Xanthomonas citri pathogenicity gene, pthA, pleiotropically encodes gratuitous avirulence on nonhosts. Mol. Plant–Microbe Interact. 5, 204–213. [DOI] [PubMed] [Google Scholar]
- Vialart, G. , Hehn, A. , Olry, A. , Ito, K. , Krieger, C. , Larbat, R. , Paris, C. , Shimizu, B. , Sugimoto, Y. , Mizutani, M. and Bourgaud, F. (2012) A 2‐oxoglutarate‐dependent dioxygenase from Ruta graveolens L. exhibits p‐coumaroyl CoA 2′‐hydroxylase activity (C2′H): a missing step in the synthesis of umbelliferone in plants. Plant J. 70, 460–470. [DOI] [PubMed] [Google Scholar]
- Wichmann, G. and Bergelson, J. (2004) Effector genes of Xanthomonas axonopodis pv. vesicatoria promote transmission and enhance other fitness traits in the field. Genetics, 166, 693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins, K.E. , Booher, N.J. , Wang, L. and Bogdanove, A.J. (2015) TAL effectors and activation of predicted host targets distinguish Asian from African strains of the rice pathogen Xanthomonas oryzae pv. oryzicola while strict conservation suggests universal importance of five TAL effectors. Front. Plant Sci. 6, 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, G.A. , Prochnik, S. , Jenkins, J. , Salse, J. , Hellsten, U. , Murat, F. , Perrier, X. , Ruiz, M. , Scalabrin, S. , Terol, J. , Takita, M.A. , Labadie, K. , Poulain, J. , Couloux, A. , Jabbari, K. , Cattonaro, F. , Del Fabbro, C. , Pinosio, S. , Zuccolo, A. , Chapman, J. , Grimwood, J. , Tadeo, F.R. , Estornell, L.H. , Muñoz‐Sanz, J.V. , Ibanez, V. , Herrero‐Ortega, A. , Aleza, P. , Pérez‐Pérez, J. , Ramón, D. , Brunel, D. , Luro, F. , Chen, C. , Farmerie, W.G. , Desany, B. , Kodira, C. , Mohiuddin, M. , Harkins, T. , Fredrikson, K. , Burns, P. , Lomsadze, A. , Borodovsky, M. , Reforgiato, G. , Freitas‐Astúa, J. , Quetier, F. , Navarro, L. , Roose, M. , Wincker, P. , Schmutz, J. , Morgante, M. , Machado, M.A. , Talon, M. , Jaillon, O. , Ollitrault, P. , Gmitter, F. and Rokhsar, D. (2014) Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 32, 656–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Q. , Chen, L.L. , Ruan, X. , Chen, D. , Zhu, A. , Chen, C. , Bertrand, D. , Jiao, W.B. , Hao, B.H. , Lyon, M.P. , Chen, J. , Gao, S. , Xing, F. , Lan, H. , Chang, J.W. , Ge, X. , Lei, Y. , Hu, Q. , Miao, Y. , Wang, L. , Xiao, S. , Biswas, M.K. , Zeng, W. , Guo, F. , Cao, H. , Yang, X. , Xu, X.W. , Cheng, Y.J. , Xu, J. , Liu, J.H. , Luo, O.J. , Tang, Z. , Guo, W.W. , Kuang, H. , Zhang, H.Y. , Roose, M.L. , Nagarajan, N. , Deng, X.X. and Ruan, Y. (2013) The draft genome of sweet orange (Citrus sinensis). Nat. Genet. 45, 59–66. [DOI] [PubMed] [Google Scholar]
- Xue, J. , Luo, D. , Xu, D. , Zeng, M. , Cui, X. , Li, L. and Huang, H. (2015) CCR1, an enzyme required for lignin biosynthesis in Arabidopsis, mediates cell proliferation exit for leaf development. Plant J. 83, 375–387. [DOI] [PubMed] [Google Scholar]
- Yan, Q. and Wang, N. (2012) High‐throughput screening and analysis of genes of Xanthomonas citri subsp. citri involved in citrus canker symptom development. Mol. Plant–Microbe Interact. 25, 69–84. [DOI] [PubMed] [Google Scholar]
- Yanagisawa, S. (1997) Dof DNA‐binding domains of plant transcription factors contribute to multiple protein–protein interactions. Eur. J. Biochem. 250, 403–410. [DOI] [PubMed] [Google Scholar]
- Yang, B. , Sugio, A. and White, F.F. (2006) Os8N3 is a host disease‐susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. USA, 103, 10 503–10 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, B. and White, F.F. (2004) Diverse members of the AvrBs3/PthA family of type III effectors are major virulence determinants in bacterial blight disease of rice. Mol. Plant–Microbe Interact. 17, 1192–1200. [DOI] [PubMed] [Google Scholar]
- Yu, Y. , Streubel, J. , Balzergue, S. , Champion, A. , Boch, J. , Koebnik, R. , Feng, J. , Verdier, V. and Szurek, B. (2011) Colonization of rice leaf blades by an African strain of Xanthomonas oryzae pv. oryzae depends on a new TAL effector that induces the rice nodulin‐3 Os11N3 gene. Mol. Plant–Microbe Interact. 24, 1102–1113. [DOI] [PubMed] [Google Scholar]
- Zhang, Z.L. , Xie, Z. , Zou, X. , Casaretto, J. , Ho, T.D. and Shen, Q.J. (2004) A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol. 134, 1500–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Peng, Z. , Long, J. , Sosso, D. , Liu, B. , Eom, J.S. , Huang, S. , Liu, S. , Vera Cruz, C. , Frommer, W.B. , White, F.F. and Yang, B. (2015) Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 82, 632–643. [DOI] [PubMed] [Google Scholar]
- Zhou, R. , Jackson, L. , Shadle, G. , Nakashima, J. , Temple, S. , Chen, F. and Dixon, R.A. (2010) Distinct cinnamoyl CoA reductases involved in parallel routes to lignin in Medicago truncatula . Proc. Natl. Acad. Sci. USA, 107, 17 803–17 808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Construction of pthA‐deletion mutants. (A) Strategy used for the deletion of the pthA genes in Xanthomonas citri strain 306. Schematic view of pthA1, pthA3 and pthA4 loci depicting the oligonucleotides (arrows) used to generate the 1.0‐kb fragments flanking the pthA coding regions (blue). The oligonucleotides PthA1F1, PthA1R2, PthA3F1, PthA3R2, PthA4F1 and PthA4R2 are specific for each pthA locus, whereas PthANR1 and PthANF2 are common to all pthAs. The bars below represent the approximate length of the DNA fragments. (B) Confirmation of the pthA deletions by polymerase chain reaction (PCR). DNAs from the single (Δ1, Δ3 and Δ4), double (Δ1‐3, Δ1‐4 and Δ3‐4) and triple (Δ1‐3‐4) mutants were amplified using the indicated oligonucleotides shown in (A) and resolved on agarose gel. The approximately 2.0‐kb PCR bands corresponding to the 1.0‐kb fragments flanking the pthA genes are indicative of the specific pthA deletions in each mutant background. (C) Examples of sequencing chromatograms of Δ1, Δ3 and Δ4 mutants showing the respective in‐frame pthA deletions. The amino acid sequences encoded by the mutated pthA loci are indicated. The 2.0‐kb fragments obtained with the specific PthAF and PthAR oligos were sequenced using oligo seqPthANF.
Table S1 Oligonucleotides used for the gene expression analyses, promoter cloning and sequencing, and pthA‐deletion mutant construction and verification.


