Summary
Plant‐parasitic nematodes cause significant damage to major crops throughout the world. The small number of genes conferring natural plant resistance and the limitations of chemical control require the development of new protective strategies. RNA interference or the inducible over‐expression of nematicidal genes provides an environment‐friendly approach to this problem. Candidate genes include NGB, which encodes a small GTP‐binding protein, and NAB/ERabp1, which encodes an auxin‐binding protein, which were identified as being up‐regulated in tomato roots in a transcriptome screen of potato cyst nematode (Globodera rostochiensis) feeding sites. Real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) and in situ hybridization confirmed the localized up‐regulation of these genes in syncytia and surrounding cells following nematode infection. Gene‐silencing constructs were introduced into tomato, resulting in a 20%–98% decrease in transcription levels. Nematode infection tests conducted on transgenic plants showed 57%–82% reduction in the number of G. rostochiensis females in vitro and 30%–46% reduction in pot trials. Transmission electron microscopy revealed a deterioration of cytoplasm, and degraded mitochondria and plastids, in syncytia induced in plants with reduced NAB/ERabp1 expression. Cytoplasm in syncytia induced in plants with low NGB expression was strongly electron translucent and contained very few ribosomes; however, mitochondria and plastids remained intact. Functional impairments in syncytial cytoplasm of silenced plants may result from NGB's role in ribosome biogenesis; this was confirmed by localization of yellow fluorescent protein (YFP)‐labelled NGB protein in nucleoli and co‐repression of NGB in plants with reduced NAB/ERabp1 expression. These results demonstrate that NGB and NAB/ERabp1 play important roles in the development of nematode‐induced syncytia.
Keywords: auxin‐binding protein, gene silencing, Globodera, GTP‐binding protein, tomato
Introduction
Plant‐parasitic nematodes are a worldwide threat to important crops. Their feeding strategies range from simple herbivory (‘hit‐and‐run’) to the formation of specialized nematode feeding sites (NFSs) within host roots. Nematodes are well equipped to locate and parasitize host plants: specialized structures, such as the stylet, enable plant tissue penetration, and oesophageal glands, secreting a cocktail of enzymes and regulators, assist nematode migration, modification of root cells into NFSs and food withdrawal (Vanholme et al., 2004). Nematode‐secreted proteins also play important roles in providing protection from the host defence response (Haegeman et al., 2012).
The majority of crop damage is caused by sedentary endoparasitic nematodes, especially the genera Meloidogyne (root‐knot nematodes), Globodera and Heterodera (cyst nematodes). Cyst nematodes usually have limited host ranges, compared with root‐knot nematodes, and cause substantial yield reduction in sugar beet (H. schachtii), potato (G. rostochiensis, G. pallida), soybean (H. glycines) and cereals (H. avenae and H. filipievi). Globodera pallida and G. rostochiensis, collectively called potato cyst nematodes (PCNs), cause serious losses in potato production, estimated at more than 12% of yield, wherever they are present (Urwin et al., 2001). Because of restrictions on nematicide use, the control of plant‐parasitic nematodes is currently achieved largely by a combination of crop rotation and the use of resistant cultivars (Gheysen et al., 1996). Natural resistance genes are highly specific to particular species, pathotypes and sometimes even races of nematode, and new virulent isolates frequently emerge which are resistant to their effects (Tomczak et al., 2009). Other sources of natural resistance in plants depend on polygene features or interaction between recessive genes (Trudgill, 1991).
The limitations in natural resistance have resulted in the development of alternative, biotechnological approaches, such as RNA interference (RNAi), to combat nematodes. These methods may be advantageous because of their broader specificity against a range of parasites. It is relatively easy to express dsRNAs of genes which modify plant development and metabolism or hamper nematode parasitism (Huang, 2006; Karczmarek et al., 2008; Lilley et al., 2012; Sindhu et al., 2009; Yadav et al., 2006). However, this technique has certain limitations, such as genus‐specific exclusion limits on the sizes of molecules, e.g. dsRNA or toxic proteins, which can be taken up by nematodes (Gheysen and Vanholme, 2007). The manipulation of the expression of genes involved in auxin signalling constitutes a promising approach for the engineering of nematode resistance because of the crucial role played by this hormone in syncytium initiation and development (Gheysen and Mitchum, 2009; Li et al., 2009). However, the pleiotropic effects of auxin can pose problems in implementing this approach. Previous research on auxin‐insensitive Arabidopsis mutants revealed the pivotal role of this hormone in the establishment of NFSs. A significant reduction in the development of beet cyst nematodes (BCNs) was observed on roots of the dominant gain‐of‐function mutant axr2, which encodes the IAA7 protein and AXR2/IAA7, a member of the auxin‐inducible Aux/IAA gene family, which controls auxin transcriptional responses (Goverse et al., 2000a). Potential auxin‐response elements were found in the promoters of genes responsible for cell wall degradation in the early stages of NFS development, suggesting the presence of an auxin‐dependent regulation mechanism (Karczmarek et al., 2008; Wang et al., 2007; Wieczorek et al., 2008). Activation of the auxin‐responsive promoter 5‐1E1 in BCN‐infected Arabidopsis roots showed that auxin accumulation may be an early marker of the reprogramming of cells about to be incorporated into the syncytium (Goverse et al., 2000b; Grunewald et al., 2009b). Furthermore, this was documented in a report describing the analyses of Arabidopsis mutants with hampered polar auxin transport (Grunewald et al., 2009a, b).
Here, we present molecular analyses of two tomato genes, NGB (NEMATODE‐INDUCED GTP‐BINDING PROTEIN) and NAB/ERabp1 (NEMATODE‐INDUCED AUXIN‐BINDING PROTEIN), known to be up‐regulated following infection of tomato roots by the PCN G. rostochiensis (Swiecicka et al., 2009). We examined mRNA expression in a variety of organs and tissues, and used RNAi in a T2 generation of stably transformed tomato plants to conduct a functional analysis, including infection tests in vitro and in pot trials. We found that changes in host susceptibility can be achieved relatively easily by modifying the expression of plant genes involved in the maintenance of NFSs. Our results have revealed the crucial and collective role played by target genes on nematode infection. Moreover, we outline their possible interaction and participation in ribosome biogenesis.
Results
Characteristics of cDNA‐AFLP candidates
Using amplified fragment length polymorphism‐based mRNA fingerprinting (cDNA‐AFLP), 17 600 transcript‐derived fragments (TDFs) were isolated from uninfected tomato roots or from roots infected with G. rostochiensis pathotype Ro1. Samples were collected 1–14 days post‐inoculation (dpi) (Swiecicka et al., 2009). Among 624 up‐regulated TDFs, 135 coding sequences were identified; two of these, NGB and NAB/ERabp1, were subjected to further detailed analysis.
NAB/ERabp1 encodes a protein similar to auxin‐binding proteins (ABPs) known from Nicotiana tabacum (88% similarity), Arabidopsis thaliana (69% similarity) and Zea mays (63% similarity; Table 1). The C‐terminal end of NAB/ERabp1 contains an endoplasmic reticulum retention signal, the KDEL motif. The protein encoded by NGB is similar to the small GTP‐binding protein of Arabidopsis thaliana (76% similarity). It has a NOG domain localized in its central part; this is characteristic of nucleolar GTP‐binding proteins. Its C‐terminal end contains an EF‐HAND‐1 domain, involved in Ca2+ binding. All of these domains are marked on the protein sequences shown in Fig. S1 (see Supporting Information).
Table 1.
Comparison of full‐length cDNA sequences with a protein database using B lastx
| Gene | Protein description and closest homologues | Similarity (%) | E value |
|---|---|---|---|
| NGB | XP004229516, nucleolar GTP‐binding protein 1‐like, Solanum lycopersicum; LOC101254633 | 99 | 0.0 |
| XP004243372, nucleolar GTP‐binding protein 1‐like, Solanum lycopersicum; LOC101249658 | 98 | 0.0 | |
| NP175505, GTP‐binding protein‐related, Arabidopsis thaliana | 76 | 0.0 | |
| NAB/ERabp1 | NP001234826, endoplasmic reticulum auxin‐binding protein 1 precursor, ERabp1, Solanum lycopersicum; | 100 | 6e‐148 |
| NP192207, ABP1‐endoplasmic reticulum auxin‐binding protein 1, Arabidopsis thaliana | 69 | 5e‐87 | |
| AAA33430, auxin‐binding protein, Zea mays | 63 | 6e‐76 | |
| AEI70327, auxin‐binding protein, Nicotiana tabacum | 88 | 6e‐117 |
Transcript levels in different organs of uninfected and nematode‐infected tomato plants
To determine the cDNA‐AFLP profiles of NGB and NAB/ERabp1, their expression levels were analysed in different organs of tomato plants using real‐time reverse transcription polymerase chain reaction (RT‐PCR). In uninfected plants, the highest levels of NAB/ERabp1 transcripts were found in leaves, whereas the highest levels of NGB mRNA (almost five times higher than in roots) were found in young fruit. Expression analysis of roots infected with G. rostochiensis (monitored at 1, 3, 7 and 14 dpi) showed maximum expression levels of NGB and NAB/ERabp1 at 7 dpi (Fig. 1).
Figure 1.
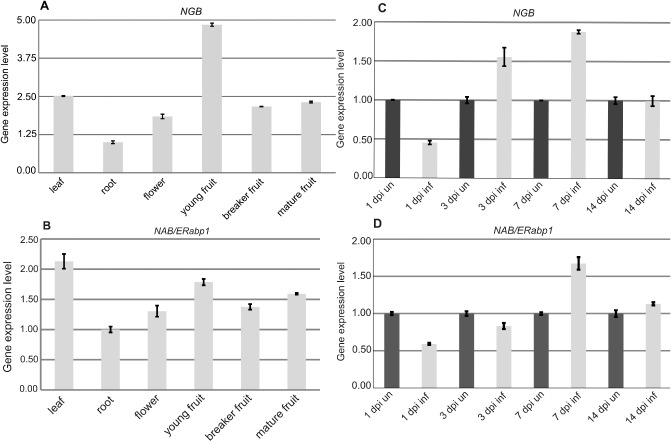
Real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of expression profiles of NGB (A, C) and NAB/ERabp1 (B, D) in different organs of uninfected tomato plants (A, B) and tomato roots infected with Globodera rostochiensis (C, D). Expression levels of NGB and NAB/ERabp1 were quantified with reference to the expression of 18S rRNA. Gene relative expression levels are shown as fold changes in comparison with the copy number of a particular mRNA in a control sample that was arbitrarily assigned a value of unity. Results presented are means (± SE) from three independent experiments.
Localization of NGB and NAB/ERabp1 expression to syncytium
We performed in situ hybridizations in sections of syncytia collected at 3, 7, 10 and 14 dpi using specific sense and antisense probes against NGB and NAB/ERabp1. NGB transcripts were detected at 3 dpi in young, expanding syncytia (Fig. 2A). During the later stages of syncytium development, the expression level of this gene was below the limit of detection (Fig. 2B). In situ hybridization of NAB/ERabp1 revealed that the transcript was present in all developmental stages of syncytium examined (Fig. 2D–I). Specific fluorescence signal appeared in both syncytial cytoplasm and in the cytoplasm of neighbouring cells. A faint fluorescence signal was observed in the oldest parts of the syncytium, localized in the cortex (cortex bridge), at 3 dpi (Fig. 2D). Later in syncytium development (7 and 10 dpi), a strong fluorescence indicating the presence of NAB/ERabp1 mRNA was observed in the cortex bridge and in vascular cylinder cells incorporated into the syncytium, as well as in cells located directly next to the syncytium, probably indicating that these cells were preconditioned to fuse with the syncytium (Fig. 2E, F). In tomato roots harbouring syncytia, expression of NAB/ERabp1 at 14 dpi had decreased in both the cortex bridge (Fig. 2G) and vascular cylinder cells (Fig. 2H), indicated by the weak fluorescence signal. In situ hybridizations with sense probes against both genes gave no signal at all tested time points (Fig. 2C, I, Fig. S7, see Supporting Information).
Figure 2.
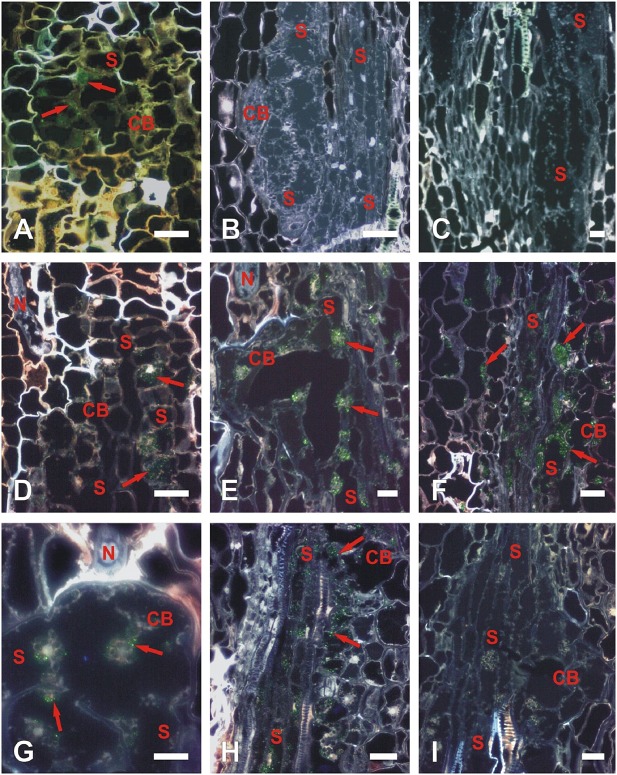
Localization of NGB (A–C) and NAB/ERabp1 (D–I) transcripts in syncytia induced by Globodera rostochiensis Ro1 pathotype in roots of wild‐type tomato plants. Localization of NGB transcripts (A–C) on cross‐sections of syncytia at 3 days post‐inoculation (dpi) (A) and 10 dpi (B, C) hybridized with antisense probe (A, B) or with sense probe as a negative control (C). Localization of NAB/ERabp1 transcripts (D–I) on cross‐sections of syncytia at 3 (D), 7 (E, I), 10 (F) and 14 dpi (G, H) hybridized with antisense probe (D–H) or with sense probe as negative control (I). Green coloration (red arrows) produced by fluorescein fluorescence indicates where antisense probes are hybridized to NGB or NAB/ERabp1 mRNA. CB, cortex bridge, a part of syncytium derived from cortex cells; N, nematode; S, syncytium. Scale bars, 20 μm.
NGB protein is localized in the nucleolus
To confirm the putative nucleolar targeting of NGB, we used NGB tagged with yellow fluorescent protein (NGB‐YFP) in a transient in vivo expression assay in Nicotiana benthamiana leaves. We observed very faint fluorescence distributed evenly across the cytoplasm and nucleoplasm, and very strong fluorescence in the nucleoli, showing that NGB is delivered to this compartment, as predicted by its bioinformatic profile (Fig. 3). The results were similar regardless of whether NGB was labelled on the N‐ or C‐terminus.
Figure 3.
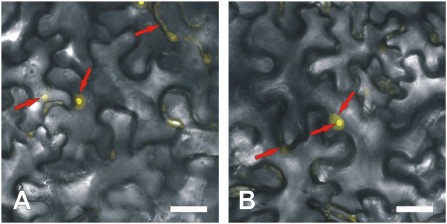
Confocal laser scanning microscopy localization of NGB‐YFP fusion protein in Nicotiana benthamiana leaf epidermis cells using Agrobacterium‐mediated transient expression. Yellow coloration (red arrows) produced by yellow fluorescent protein (YFP) fluorescence (artificial colour) indicates sites at which fusion protein has accumulated in the cytoplasm (faint colour), nucleoplasm (moderate colour intensity) and nucleolus (strong yellow coloration). NGB was labelled on the N‐ (A) or C‐ (B) terminus. Scale bars, 20 μm.
Tomato transformation and analysis
The ability of constructs to reduce the expression of target genes was first checked by transiently expressing the uidA reporter gene and silencing constructs in N. benthamiana leaves (Wroblewski et al., 2005). The presence of inversely repeated fragments of the reporter gene fragments in silencing constructs allowed the checking of whether or not the vector functioned correctly before performing time‐consuming stable tomato transformation (Figs S2 and S3, see Supporting Information). Subsequently, in stable transformation experiments, we obtained 10 independent fertile transformed tomato lines containing the NGB‐silencing construct and 10 lines containing the NAB/ERabp1‐silencing construct. Thus, the transformation efficiency (percentage of independently rooted plants per explant number) differed for each of the silencing constructs; the transformation efficiency was higher for NAB/ERabp1 (3.3%) and lower for NGB (1.7%; Table S1, see Supporting Information). Genetic analysis revealed that nine (of 10) fertile transgenic tomato lines containing the NGB‐silencing construct had a single transgene integration site in the genome; this was the case for seven (of 10) fertile lines containing the NAB/ERabp1‐silencing construct. Homozygous T1 tomato lines were subsequently selected from single‐copy transformants by analysing the segregation of kanamycin resistance in T2 progeny (Table S2, see Supporting Information). The presence of NGB‐ and NAB/ERabp1‐silencing constructs in selected homozygous T1 plants was verified by PCR with primers specific for the reporter gene, uidA, and the kanamycin selection marker, nptII, also present in the silencing constructs (Fig. S4, see Supporting Information).
Real‐time RT‐PCR was used to evaluate the level of reduction of target gene expression in homozygous transgenic lines (Fig. 4). In six homozygous lines containing the NGB‐silencing construct, the reduction in NGB expression ranged from 80% to 90% compared with control plants. Moderate levels of silencing of NGB mRNA (20%–60%, relative to wild‐type plants) were also observed. In addition, three homozygous transgenic lines containing the silencing construct over‐expressed the target gene (Fig. 4). A reduction of 95%–98% in NAB/ERabp1 expression level was achieved in homozygous lines containing the NAB/ERabp1‐silencing construct, compared with control plants, although some lines showed a lower reduction in expression level (70%–85%; Fig. 4). In each case, the effect on gene expression was stable across generations, as confirmed by real‐time RT‐PCR analysis of expression in T2 plants (Fig. S5, Tables S3 and S7, see Supporting Information).
Figure 4.
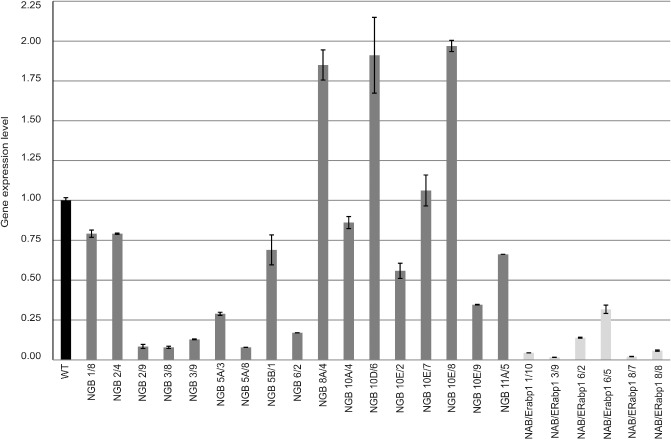
Real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of NGB and NAB/ERabp1 expression levels in leaves of T1 homozygous tomato plants transformed with silencing constructs. Expression levels of NGB and NAB/ERabp1 were quantified with reference to the expression of 18S rRNA. Relative expression levels are shown as fold changes relative to the copy number of a particular gene mRNA in a control sample that was arbitrarily assigned a value of unity. Results are means (± SE) from three independent experiments.
Reduction in NAB/ERabp1 and NGB expression results in reduced susceptibility to G. rostochiensis
Homozygous lines with the greatest reduction in NAB/ERabp1 or NGB expression were used in further in vitro infection tests. A decrease in the number of infection sites on roots of transgenic plants was observed at 7 dpi in all transgenic lines, relative to control plants, although, at 14 dpi, new infection sites were observed in all control and transgenic lines. Groups of necrotized root cells delineate the migration paths of second‐stage infective juveniles. Degraded and well‐developed syncytia were observed in both transgenic and control lines; however, at the end of the experiment (42 dpi), the number of fully developed females was 57%–82% lower on transgenic tomato roots than on controls (Fig. 5). The same set of transgenic plant lines was subjected, in pot trials, to infection tests with G. rostochiensis pathotype Ro1 (Fig. S8, see Supporting Information). Preliminary analyses showed a reduction of 30%–46% in the number of females, compared with the number on control plants, in most genotypes examined; the exception was transgenic line NGB 5A/8, which appeared to be more susceptible than control plants.
Figure 5.
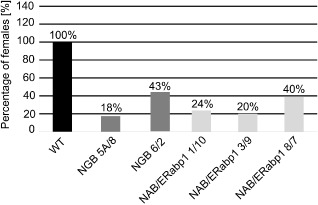
Relative susceptibility of silenced transgenic homozygous tomato lines to Globodera rostochiensis pathotype Ro1 in vitro. Values are expressed as a percentage of the number of females which developed on transgenic tomato roots relative to the number on control roots. Number of females on control roots was assigned a value of 100%.
Reducing the expression of NGB or NAB/ERabp1 disturbs the expression of other genes
Although silencing constructs were driven by the CaMV35S promoter, often defined as a constitutive regulatory element, we observed that the effects of reducing the levels of NAB/ERabp1 or NGB on the expression of other genes varied with respect to the plant organ and individual transformation event. Generally, effective reduction of NAB/ERabp1 expression in roots had a clear effect on auxin‐response genes, resulting in significant over‐expression of tomato AUXIN‐RESPONSE FACTOR 16 (ARF16; Solyc07g00802) and IAA AMIDOSYNTHETASE (Solyc02g064830); however, in above‐ground parts of the plant, this effect was insignificant. Instead, we observed an induction in transcripts coding for the pathogenesis‐related proteins PR1 and PR6 in above‐ground organs. Similarly, reductions in NGB expression induced a two‐fold increase in the expression of ARF16 and IAA in the roots of transgenic plants, relative to wild‐type controls, accompanied by a similar induction of PR genes. In leaves, increased induction was observed only for PR6 (Fig. 6A, B).
Figure 6.
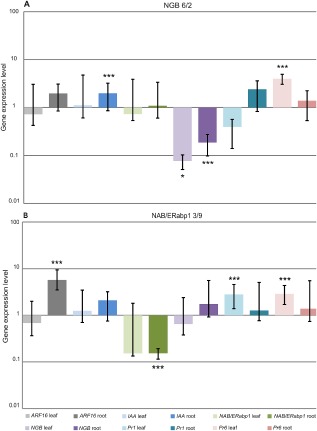
Real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of auxin markers and pathogenesis‐related gene expression in leaves and roots of homozygous transgenic tomato lines NGB 6/2 (A) and NAB/ERabp1 3/9 (B). Expression levels of target genes were quantified with reference to the expression of SAND and LPE8. Relative expression levels are shown as fold changes relative to the copy number of a particular gene mRNA in a control sample. Results are means (± SE) from three independent experiments. The asterisks indicate the significance of the differences from the control as revealed by REST (Pfaffl et al., 2002): ***P < 0.001; *P < 0.05.
Down‐regulation of NGB or NAB/ERabp1 disturbs syncytia development
Infective second‐stage juveniles of G. rostochiensis Ro1 pathotype were able to invade roots of T2 tomato plants with greatly reduced expression of NGB or NAB/ERabp1 (Fig. 5). Light microscopy examination revealed that syncytia in infected transgenic plants followed a developmental pathway typical for G. rostochiensis‐induced syncytia in tomato (Sobczak et al., 2005). Syncytia were first induced among cortical cells and spread towards the vascular cylinder through the incorporation of cortical and endodermal cells into the cortex bridge, which formed next to the juvenile head (Fig. 7A, C, G, I, K, M, O, Q, S, W). On reaching the vascular cylinder, the syncytium incorporated pericyclic cells and then expanded between xylem and phloem bundles by recruiting procambial and cambial cells (Fig. 7A, C, E, G, I, K, M, O, Q, S, U, W). The distal parts of the syncytia were composed of elements derived from the vascular cylinder cells only (Fig. 7E, U).
Figure 7.
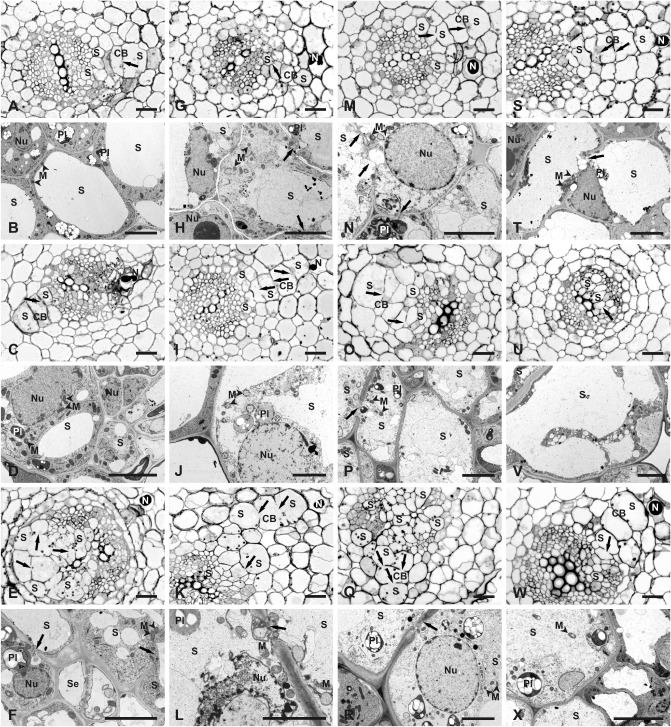
Comparison of anatomy and ultrastructure of syncytia induced by Globodera rostochiensis in roots of wild‐type and transgenic plants. Light microscopy (A, C, E, G, I, K, M, O, Q, S, U and W) and transmission electron microscopy (B, D, F, H, J, L, N, P, R, T, V and X) images of cross‐sections of syncytia induced in roots of the wild‐type cultivar ‘MoneyMaker’ (A–F) or transgenic plants containing silencing constructs (G–X): syncytia in transgenic line 3/9 containing NAB/ERabp1‐silencing construct (G–L); syncytia in transgenic lines 6/2 (M–R) and 5A/8 (S–X) containing NGB‐silencing construct. Syncytia were examined at 4 days post‐inoculation (dpi) (A, B, G, H, M, N, S and T), 7 dpi (C, D, I, J, O, P, U and V) and 14 dpi (E, F, K, L, Q, R, W and X). CB, cortex bridge; M, mitochondrion (indicated by neighbouring arrowheads); N, nematode; Nu, nucleus; Pl, plastid; S, syncytium. Arrows point to cell wall openings. Scale bars, 20 μm (A, C, E, G, I, K, M, O, Q, S, U and W); 5 μm (B, D, F, H, J, L, N, P, R, T, V and X).
Transmission electron microscopy (TEM) examination revealed several differences in ultrastructure between syncytia induced in roots of the susceptible tomato cultivar ‘MoneyMaker’ (Fig. 7B, D, F) and in T2 plants with greatly reduced expression of NAB/ERabp1 (Fig. 7H, J, L) or NGB (Fig. 7N, P, R, T, V, X). The cytoplasm was usually less electron opaque in syncytia induced in roots of plants bearing the NAB/ERabp1‐silencing construct (Fig. 7H) than in control plants (Fig. 7B), and they also contained fewer endoplasmic reticulum structures; however, other aspects of their ultrastructure (a decrease in central vacuole size, intact plastids and mitochondria, and hypertrophy of the nucleus) resembled typical syncytia at 4 dpi. Beyond 7 dpi, almost all examined syncytia contained granular and plasmolysed cytoplasm with degraded plastids, mitochondria and nuclei (Fig. 7J, L). A different set of ultrastructural features was observed in syncytia induced in the roots of plants containing the NGB‐silencing construct. Such syncytia usually contained flocculent and strongly electron‐translucent cytoplasm at 4 dpi (Fig. 7N, T) and electron‐translucent cytoplasm was also present in syncytia examined at 7 and 14 dpi (Fig. 7P, R, V, X). Comparison at high magnification of syncytial cytoplasm and cytoplasm of neighbouring cells indicated that very few ribosomes were present in the former (Fig. S6, see Supporting Information). As with plants containing the NAB/ERabp1‐silencing construct, only a few endoplasmic reticulum structures were present in the syncytial cytoplasm at all time points; by contrast, however, no degeneration of the syncytial cytoplasm occurred (Fig. 7P, R, V, X). In addition, the ultrastructure of mitochondria and plastids remained intact and showed no features of degradation, even in syncytia collected at 14 dpi (Fig. 7R, X). In transgenic plants containing NAB/ERabp1‐silencing constructs, nuclei of syncytia acquired regular round outlines on section and their nucleoplasm was strongly electron translucent (Fig. 7J), similar to the observations in syncytia induced in plants containing the NGB‐silencing constructs (Fig. 7N, R). By contrast, nuclei of syncytia in control plants were amoeboid in shape and osmiophilic (Fig. 7D). However, nuclear envelope rupture or nucleus collapse occurred in syncytia induced in plants with very low expression of NAB/ERabp1 (Fig. 7L), but not in syncytia in plants with low NGB expression (Fig. 7N, R).
Discussion
Research into genes involved in the plant–nematode interaction may aid in the development of effective pest management strategies. In particular, the development of strategies based on RNAi inhibition of target genes crucial for pathogenesis has produced promising results (Karczmarek et al., 2008; Lilley et al., 2012). Here, we examined the effects of the suppression of NGB and NAB/ERabp1 expression on the interaction of tomato plants with PCN. Both genes were isolated during transcriptome profiling of tomato roots infected with PCN (Swiecicka et al., 2009). The sequence similarity and subcellular localization of NGB point to its role in ribosome biogenesis. The association of ribosomal biogenesis with deficient auxin perception and distribution has been widely reported in the literature (e.g. Rosado et al., 2010). For example, mutation in the nucleolar protein PARL1 involved in ribosome synthesis causes auxin‐related developmental defects, suggesting that auxin regulation depends on protein turnover and ribosome biogenesis (Petricka and Nelson, 2007). In addition, the closest NGB homologue in Arabidopsis, Nog1‐1, plays a critical role in plant development, as evidenced by the quite severe growth impairment of Nog1‐1 RNAi‐silenced plants (Suwastika et al., 2008).
Characteristics of NGB and NAB/ERabp1 genes and proteins
NGB shows similarity to small nuclear GTP‐binding proteins containing the NOG domain, which can be found in a wide range of eukaryotic and prokaryotic species. Its homologue, NOG1, in Trypanosoma brucei is functionally linked to pre‐ribosomal complexes made up of the 60S subunit and nuclear pore complexes (Fuentes et al., 2007; Jensen et al., 2003). The NOG domain is also characteristic of small G proteins of the Ras protein superfamily, whose members are involved in signal transduction and regulation of gene expression (Assmann, 2002, 2005). The C‐terminus of NGB contains an EF‐HAND‐1 domain which may be involved in Ca2+ binding, a universal secondary messenger involved in plant growth and development, as well as in the mediation of responses to biotic and abiotic stress (Anderson and Botella, 2007). NAB/ERabp1 is similar to an ABP purified from maize coleoptiles (Hesse et al., 1989; Palme et al., 1990). It contains a cleavable N‐terminal signal sequence and a C‐terminal signal element, consisting of the amino acids lysine‐aspartic acid‐glutamic acid‐leucine (Lys‐Asp‐Glu‐Leu), which are responsible for retaining the protein in the endoplasmic reticulum lumen. It has been suggested that ABPs could be involved in the regulation of auxin‐induced cell elongation (Jones, 1994), play a role in cell cycle control and be key regulators of root growth (David et al., 2007; Perrot‐Rechenmann, 2010; Tromas et al., 2009) and stomatal closure (Montillet et al., 2013). The involvement of auxin in syncytium formation has been mainly considered in the context of cell division regulation and cell expansion (Goverse et al., 2000a, b; Wang et al., 2007). Goverse et al. (2000b) analysed the auxin‐insensitive Arabidopsis and tomato mutants dgt and axr2, which revealed a pivotal role of this hormone in the early stages of syncytium development. Wide analyses of Arabidopsis mutants with perturbations in polar auxin transport and infected with BCN showed that both acropetal auxin transport and induced translocation of auxin in young feeding cells are required for syncytium development (Grunewald et al., 2009a). Moreover, in silico analyses of nematode‐responsive genes showed that many of them contain auxin‐response elements, suggesting that transcriptional activation occurs via the accumulation of auxin on nematode infection (Swiecicka et al., 2009; Wang et al., 2007; Wieczorek et al., 2008). This hypothesis is supported by analyses of the auxin‐repressed genes ADR6, ADR11 and ADR12, which are differentially expressed in soybean roots on infection with the soybean cyst nematode H. glycines (Hermsmeier et al., 1998). However, analysis of auxin‐inducible WRKY23 gene suggested that auxin‐independent signals may be involved in the activation of their expression in the early stage of NFS development (Grunewald et al., 2008).
Expression analysis of NGB and NAB/ERabp1 genes
We have reported previously that NGB and NAB/ERabp1 are both significantly up‐regulated during syncytium development (Swiecicka et al., 2009). This indicates that they may play important roles in signal transduction events induced by G. rostochiensis parasitism. Expression of NAB/ERabp1 in tomato clearly correlated with intensively growing tissues but, in contrast with the abp1 mutant of Arabidopsis (Effendi et al., 2011), we did not observe any lethal effects resulting from the near‐total silencing of this gene. We visualized the locations of NAB/ERabp1expression in situ across all stages of syncytium development (1–14 dpi), and observed mRNA expression in both syncytia and surrounding cells. We suggest that NAB/ERabp1 is involved not only in the induction and expansion of syncytia, but also in their continued maintenance, as TEM observations indicate that young syncytia induced in plants with suppressed NAB/ERabp1 expression initially have an ultrastructure similar to that induced in wild‐type plants, but subsequently degenerate, indicating a loss of morphogenetic control. The spatial distribution of NGB expression was also assessed using promoter‐β‐glucuronidase (promoter‐GUS) fusions. GUS expression was observed in lateral root primordia, apical meristems of main and lateral roots, and in developing syncytia (A. Wiśniewska, unpublished data). Taken in conjunction with our in situ hybridization data, we conclude that NGB is expressed specifically in tissues undergoing intensive cell division during the early stages of syncytium expansion (Golinowski et al., 1996).
The role of NGB and NAB/ERabp1 genes in cyst nematode parasitism
NAB/ERabp1 and NGB apparently play pleiotropic roles in tomato and participate in distinct processes in different organs. Post‐transcriptional silencing of NAB/ERabp1 or NGB resulted in a decrease in the number of female nematodes on roots, probably because prematurely degraded syncytia cannot support the development of juveniles. The changes in syncytia ultrastructure induced in plants containing NAB/ERabp1‐silencing constructs resemble those induced by plant defence reactions in tomato plants carrying HeroA or Cf‐2 resistance genes (Sobczak et al., 2005; Lozano‐Torres et al., 2013) or potato plants carrying the H1 resistance gene (Rice et al., 1985). However, the additional changes we observed in syncytia induced in plants with very low NGB expression, including an electron‐translucent cytoplasm containing fewer ribosomes and structurally unchanged organelles, are exceptional and have not been described previously. Kandoth et al. (2011) indicated the possibility of a relationship between soybean Rhg1‐dependent cyst nematode resistance and the unfolded protein response. The ribosome biogenesis disturbances postulated in NGB‐silenced plants may contribute to a similar resistance mechanism. Our data also indicate that relatively small changes in the cellular auxin equilibrium may be sufficient to disturb syncytium development and suggest the involvement of auxin‐binding proteins. We observed changes in the expression of auxin and pathogenesis‐related genes in NAB/ERabp1‐suppressed plants; such pleiotropic effects caused by a decrease in NAB/ERabp1 expression are expected because of its involvement in a large number of cellular processes. The induction of auxin‐dependent and pathogenesis‐related genes in NAB/ERabp1‐suppressed plants may explain their higher nematode resistance, longer roots (Fig. S9, see Supporting Information) and syncytium degeneration. Little is known about the functioning of small GTP‐binding proteins in plants during pathogenesis. However, the β‐subunit of the heterotrimeric G protein AGB1, which is involved in the control of organ shape in Arabidopsis, is required for full disease resistance to Pseudomonas syringae strains (Torres et al., 2013). Furthermore, some pathogenesis‐related G proteins, including AGB1 and AGG1/AGG2, function downstream of multiple receptor‐like kinases that activate resistance responses to pathogens (Liu et al., 2013). Differential regulation of PR1 and PR6 in NGB‐suppressed plants may be a consequence of its involvement in such pathways. The expression analysis of NGB and NAB/ERabp1 in the present study indicates co‐regulation of these genes. The auxin‐related effects of their silencing, i.e. auxin marker gene up‐regulation and changes in root growth, indicate the presence of a coordinated mechanism of action which may depend on protein turnover and ribosome biogenesis. We speculate that auxin acts via G proteins to regulate gene expression and, reciprocally, auxin regulation depends on ribosome biogenesis regulated by NGB. The disturbance of this type of regulatory homeostasis in our transgenic lines, where target genes were not fully silenced, may explain the syncytium degeneration observed here. Metabolically less demanding conditions during the normal growth of tomato plants may not be sufficient to cause stronger effects.
Experimental Procedures
Bioinformatics analyses of nucleotide and protein sequences
The NGB gene was identified by reference to cDNA sequences deposited in expressed sequence tag (EST) databases: the Gene Indices database (http://compbio.dfci.harvard.edu/tgi/) and GenBank dbEST (http://www.ncbi.nlm.nih.gov/genbank/dbest/). The full‐length cDNA sequence (containing the entire open reading frame) was reconstructed manually. The NAB/ERabp1 sequence was obtained from the GenBank database. Using the Blastx algorithm (Altschul et al., 1990), we identified sequences similar to proteins in the databases of non‐redundant protein sequences, using the BioEdit package (Sequence Alignment Editor; http://www.mbio.ncsu.edu/bioedit/bioedit.html) to predict putative protein sequences. Conserved protein motifs were identified using the Conserved Domain Database (CDD; http://www.ncbi.nlm.nih.gov; Marchler‐Bauer et al., 2011), PROSITE (http://us.expasy.org/prosite/; Sigrist et al., 2002) and SMART (http://smart.embl‐heidelberg.de; Letunic et al., 2012). Protein schematics were visualized using the dog program (http://dog.biocuckoo.org/; Ren et al., 2009).
Real‐time RT‐PCR
Total RNA was isolated from flowers, leaves and young, breaker and mature fruits of tomato plants (Solanum lycopersicum). Samples were collected from wild‐type and transgenic tomato plants whose roots were infected with PCNs (Globodera rostochiensis, Ro1 pathotype) and from uninfected controls.
Sequences of gene‐specific primers and reaction parameters are shown in Table S3. RNA was isolated using Trizol® (Invitrogen™/Life Technologies™, Frankfurt, Germany), according to the manufacturer's protocol for samples rich in polysaccharides. Before RT‐PCR, RNA was treated with RNase‐free DNase I (Fermentas/Thermo Scientific, Carlsbad, CA, USA) to eliminate contamination from genomic DNA. RNA purity was checked spectrophotometrically (Beckman DU‐70, Los Angeles, CA, USA) by measuring the absorbance at 260 and 280 nm, and RNA quality was checked electrophoretically on 1% agarose gels (Prona, Madrid, Spain) with ethidium bromide (Sigma, St. Louis, MO, USA) staining.
The relative expression of selected genes was assessed using real‐time RT‐PCR analysis and expressed as the number of copies of the amplified tested gene per copy of the endogenous control gene 18S rRNA. Real‐time RT‐PCR was performed using the LightCycler RNA Amplification Kit and SYBR Green I (Roche, Basle, Switzerland), according to the manufacturer's instructions. Three independent experiments and three technical repeats were performed. The reaction mixture composition and reaction parameters are shown in Tables S5 and S7 (see Supporting Information).
Total RNA from roots and leaves of 14‐day‐old tomato plants from transgenic lines NAB/ERabp1 3/9 and NGB 6/2 was isolated according to the protocol of Chomczynski (1993). Gene‐specific primers and reaction parameters are shown in Table S4 (see Supporting Information). Before RT‐PCR, RNA was treated with RNase‐free DNase I (Fermentas/Thermo Scientific) to eliminate contamination from genomic DNA. RNA purity was checked spectrophotometrically (Nanodrop 1000 spectrophotometer, Thermo Scientific) by measuring the absorbance at 260 and 280 nm. The quality of RNA was checked electrophoretically on 1% agarose gels (Prona) with ethidium bromide (Sigma) staining. The relative expression of selected genes was assessed using real‐time RT‐PCR analysis, and expressed as the number of copies of the amplified tested gene per copy of the endogenous control genes RPL8 and Sand (Expósito‐Rodríguez et al., 2008). Real‐time RT‐PCR was performed with the Bio‐Rad CFX96 Touch™ Real‐Time PCR Detection System (Bio‐Rad, Niasto, CA, USA) and the iScript™ One‐Step RT‐PCR kit with SYBR® Green (Bio‐Rad), according to the manufacturer's instructions. Three independent experiments and three technical repeats were performed. The significance of differences from the control was revealed by REST (Pfaffl et al., 2002). The reaction mixture composition and reaction parameters are shown in Tables S6 and S8 (see Supporting Information).
In situ hybridization
Wild‐type tomato plants, cultivar ‘MoneyMaker’, were grown on agar‐solidified 0.5 × Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) for 2 weeks. They were inoculated, under sterile conditions, with freshly hatched surface‐sterilized infective second‐stage juveniles of G. rostochiensis, Ro1 pathotype. Root samples containing syncytia were collected at 3, 7, 10 and 14 dpi, and processed as described by Fudali et al. (2008) and Karczmarek et al. (2008).
Single‐stranded cDNA probes for in situ hybridization with NAB/ERabp1 and NGB mRNA were synthesized in asymmetric PCR using the following primer pairs: 5′‐CTCGAAACGACGGTACGGTACGGTGATG‐3′ and 5′‐GCCTGTGGATTGGTGTGCGACAT‐3′ for NAB/ERabp1 and 5′‐CAACAGAATTTTGGGTGACTACTAGA‐3′ and 5′‐ACGATAAAATAAACGGTAGCAAACT‐3′ for NGB. The prehybridization, hybridization and detection steps were performed according to the procedures described by Fudali et al. (2008) and Karczmarek et al. (2008).
Sections were examined with an Olympus AX70 ‘Provis’ (Olympus, Tokyo, Japan) fluorescence microscope equipped with a U‐M61002 triple band filter set and an Olympus DP50 digital camera. All images were equilibrated for similar contrast and brightness using Adobe Photoshop software.
Preparation of silencing constructs
Silencing constructs were made using the pDraCULA vector (Wroblewski et al., 2007) (Fig. S2). Short inverted repeat sequences from the genes to be silenced were introduced into this vector in a single step using SfiI restriction sites. The sequence recognized by this enzyme makes it possible to clone gene fragments in a specific orientation relative to the PDK intron (Flaveria trinervia pyruvate dikinase; accession no. X79095). Short sequences from NGB were amplified using the primers 5′‐ACAGAATTTTGGGTGACTACTAGA‐3′ and 5′‐AGAGACAGCAAATTCAACTACACA‐3′ (138 bp); short sequences from NAB/ERabp1 were amplified using the primers 5′‐TCGAAACGACGGTACGGTG‐3′ and 5′‐TGTGGATTGGTGTG‐3′ (303 bp). The SfiI restriction site, 5′‐GGCCTGAGTGGCC‐3′, was added to the 5′ end of all primers.
Gene amplification was performed using the BD Advantage™ 2 PCR Enzyme System (BD Biosciences‐Clontech, Mountain View, CA, USA). Restriction analysis of inserts and modification of vector ends were performed according to the manufacturer's instructions (Fermentas/Thermo Scientific). Ligation reactions were performed using Ready‐to‐go T4 DNA ligase from bacteriophage T4 (Ready‐to‐go T4 DNA Ligase, Amersham/GE Healthcare Life Sciences, Piscataway, NJ, USA). Fragments for ligation were recovered from 1% agarose gels and purified using a QIAquickR Gel Extraction Kit (Qiagen, Hilden, Germany). Silencing constructs were introduced into Agrobacterium tumefaciens C58C1 using electroporation, according to the Escherichia coli Pulser manual (Bio‐Rad).
Transient expression of silencing constructs in N. benthamiana leaves
The silencing activity of NGB and NAB/ERabp1 constructs in pDraCULA vector was checked in Nicotiana benthamiana leaves using a transient transformation assay (Wroblewski et al., 2005; Fig. S3). An individual A. tumefaciens colony, grown on solid YEB medium (5 g/L beef extract, 1 g/L yeast extract, 5 g/L peptone, 5 g/L sucrose, 0.5 g/L MgCL2, 1.5 % agar) supplemented with selective antibiotic, was cultured overnight in liquid YEB medium without selective agent at a temperature of 28 °C, 180 rpm agitation. Overnight cultures were transferred to fresh liquid YEB medium in a 1:10 ratio and incubated at a temperature of 28 °C, 180 rpm agitation, for 7 h. Afterwards, bacterial cultures were centrifuged for 5 min at 1000 g and resuspended in MilliQ (Merck, Warsaw, Poland) water [optical density at 600 nm (OD600) = 0.4–0.5].
N. benthamiana plants were grown in a growth chamber under controlled conditions (light : dark photoperiod, 16 h : 8 h; warm : cold thermoperiod, 25 °C : 18 °C; light intensity, 54 μmol/m2/s). Young (second, third and fourth) leaves were syringe infiltrated with a mixture of A. tumefaciens transformed with the silencing construct and a functional reporter construct, 35S::uidA, or with bacteria containing only the reporter construct (positive control). Leaves were dissected after 3 days and placed in 0.05 m phosphate buffer (pH 7.2) containing 0.1% 5‐bromo‐4‐chloro‐3‐indolyl beta‐D‐glucuronide (X‐GlcA) (Duchefa, Haarlem, the Netherlands), 20% methanol and 0.1% Triton X‐100 (Sigma). They were incubated at 37 °C for 24 h, and then the reaction was stopped with 100% ethanol. Leaves were washed several times with ethanol until complete removal of chlorophyll. uidA gene activity was determined on the basis of visual assessment of colour intensity.
Tomato transformation
NGB and NAB/ERabp1 silencing constructs were introduced into the tomato genome by stable A. tumefaciens‐mediated transformation (Frary and Earle, 1996).
Nematode inoculation
Seeds of transgenic and control tomato plants were sterilized using 70% ethanol (POCH, Gliwice, Poland) (1–2 min), sodium hypochlorite (POCH) (15 min) and washed three times in sterile water: 5, 10 and 15 min, respectively. After sterilization, seeds were transferred to a 0.8% agar Petri dish and incubated for 3 days in a dark growth chamber. Then, germinated seeds were grown on agar‐solidified 0.5 × MS medium (Murashige and Skoog, 1962), pH 6.2, for 2 weeks under controlled conditions (light : dark photoperiod, 16 h : 8 h; warm : cold thermoperiod, 25 °C : 18 °C; light intensity, 54 μmol/m2/s). Each 0.5 × MS Petri dish contained one tomato seedling. After 2 weeks, above‐ground plant parts were cut off and sterile tomato roots were inoculated with 200 freshly hatched, sterile, second‐stage juveniles of G. rostochiensis Woll. pathotype Ro1 per root system. The juveniles were hatched from dry cysts, as described by Goverse et al. (2000b). Altogether, 20 root systems for each tested transgenic tomato line and control plants were inoculated in two experiments. Afterwards, plates were transferred to a dark growth chamber and incubated at a temperature of 18 °C. Observations were performed at 7, 14, 21 and 42 dpi using a binocular microscope (Olympus).
G. rostochiensis pathotype Ro1 was also used in pot trials. Seeds of transgenic and control tomato plants were incubated in water for 1 h and transferred to a Petri dish with wet blotting paper before being incubated for 3 days at a temperature of 22 °C. Germinating seeds were transferred to small pots filled with a sterile sand‐soil mixture (1:4) and placed in a glasshouse. After 2 weeks, plants were transferred to 1‐L pots filled with a sterile sand‐soil mixture containing cysts of G. rostochiensis. This provided an initial population density of 10 viable eggs/mL soil (six plants for each tested transgenic tomato line and control plants). Plants were grown in the glasshouse under controlled conditions (light : dark photoperiod, 16 h : 8 h; humidity, 59%). The numbers of females that developed on transgenic and control plant roots were counted after 3.5 months of culture.
Light microscopy and TEM
Wild‐type and transgenic plants were grown and inoculated as described above for collection of samples for in situ hybridization. Samples were collected at 4, 7 and 14 dpi. They were immediately fixed in a mixture of paraformaldehyde and glutaraldehyde, and processed for microscopic examination (Sobczak et al., 2005). Semi‐thin sections were examined with an Olympus AX70 ‘Provis’ light microscope equipped with an Olympus DP50 digital camera. Ultrathin sections were examined with an FEI 268D ‘Morgagni’ transmission electron microscope (FEI Comp., Hillsboro, OR, USA), operating at 80 kV and equipped with a 10MPix digital camera ‘Morada’ (Olympus‐SIS, Münster, Germany). All rough images were equilibrated for similar contrast and brightness using Adobe Photoshop software.
Supporting information
Fig. S1 Schematic diagram of predicted protein sequences encoded by NAB/ERabp1 (A) and NGB (B). The NAB/ERabp1 protein sequence possesses a highly conserved KDEL motif, a signal of endoplasmic reticulum localization, near the C‐terminus (A). The NGB protein sequence contains a NOG domain in its central part. This is characteristic of low‐molecular‐weight GTP‐binding proteins belonging to the Ras superfamily. NOGCT is a NOG C‐terminal domain. An EF‐HAND‐1 domain (EF‐hand calcium‐binding domain) involved in Ca2+ binding is located near the protein's C‐terminus (B). Both domains are activators of several proteins involved in cell‐to‐cell signal transduction.
Fig. S2 Architecture of silencing constructs in pDraCULA (Wroblewski et al., 2007). The pDraCULA vector contains the inverted repeats of a reporter gene, uidA, in order to determine the correct function of the silencing constructs. Transcription of the hairpin structure (dsRNA) is controlled by the CaMV35S promoter and a polyadenylation signal from Agrobacterium tumefaciens octopine synthase (OCS). The selection marker, nptII, conferring kanamycin resistance, is controlled by the A. tumefaciens nopaline synthase (NOS) promoter and polyadenylation signal present in the T‐DNA region. The lengths of the target gene silencing fragments were 138 bp (NGB) and 303 bp (NAB/ERabp1).
Fig. S3 Transient expression of silencing constructs in Nicotiana benthamiana leaves. Tobacco leaves infiltrated with Agrobacterium tumefaciens C58C1 strain containing the reporter construct 35S::uidA (left side of each leaf) or the reporter construct 35S::uidA (right side of each leaf) and a gene‐silencing construct pDraCULA::NAB/ERabp1SfiI (A) or pDraCULA::NGBSfiI (B). Scale bars, 1.5 cm. The silencing activity of NGB and NAB/ERabp1 gene constructs in pDraCULA was checked in N. benthamiana leaves in a transient transformation assay (Wroblewski et al., 2005). The inclusion of inversely repeated fragments of the reporter gene uidA allows the verification of correct vector functioning before stable plant transformation, as the target genes were driven by the same constitutive CaMV35S promoter. The transient assay was carried out by co‐infiltration of N. benthamiana leaves with A. tumefaciens strains harbouring the silencing constructs and monitoring of uidA gene activity.
Fig. S4 Polymerase chain reaction (PCR) results confirming the presence of transgenes in homozygous T1 tomato plants. PCR was carried out using gene‐specific primer pairs for uidA and nptII to confirm the presence of the uidA reporter gene in sense (A) and antisense (B) orientations, and the nptII gene (C) in the genomes of selected transgenic tomato plants transformed with NGB or NAB/ERabp1 gene‐silencing constructs. Fragments of the expected length (300 bp for uidA and 700 bp for nptII) were amplified and PCR products were analysed using gel electrophoresis. Abbreviations: M, DNA marker (SMO331, MBI Fermentas); N, genomic DNA from wild‐type tomato plant (negative control); P1, plasmid DNA 35S::NGB/pDraCULA (positive control); P2, plasmid DNA 35S::NAB/ERabp1/pDraCULA (positive control); 1, 2, genomic DNA of transgenic tomato plants containing the NGB‐silencing construct (lines: 1, NGB 6/2; 2, NGB 5A/8); 3–5, genomic DNA of transgenic tomato plants with the NAB/ERabp1‐silencing construct (lines: 3, NAB/ERabp1 1/10; 4, NAB/ERabp1 3/9; 5, NAB/ERabp1 8/7). The primer sequences used for the amplification of uidA and nptII genes were as follows: nptIIF, 5′‐GAGGCTATTCGGCTATGACTG‐3′; nptIIR, 5′‐ATCGGGAGCGGCGATACCGTA‐3′; uidAF, 5′‐TACACCACGCCGAACACCTG‐3′; uidAR, 5′‐AAACAAATGCCTAAAGAGAG‐3′. Primer annealing temperature, 55 °C; elongation time, 30 s for uidA and 60 s for nptII.
Fig. S5 Expression levels of NGB and NAB/ERabp1 in leaves of homozygous T2 tomato plants carrying gene‐silencing constructs. The silencing effect of the constructs on target gene expression in homozygous T2 tomato lines was determined by real‐time reverse transcription‐polymerase chain reaction (RT‐PCR); expression levels of NGB and NAB/ERabp1 were quantified with reference to the expression of 18S rRNA. The stability of gene silencing was confirmed in transgenic lines selected for further examination. In homozygous T2 plants, we observed a reduction in NGB expression of 76% in NGB lines 5A/8 and 6/2, a reduction of 37% and in NAB/ERabp1 lines 3/9, 8/7 and 1/10, a reduction in NAB/ERabp1 expression of 78%, 88% and 73%, respectively. Relative expression levels are shown as fold changes in comparison with the expression of a given mRNA in a control sample that was arbitrarily assigned a value of unity. The results presented are means (± SE) obtained from three independent experiments.
Fig. S6 Comparison of cytoplasm ultrastructure and ribosomal abundance between neighbouring non‐syncytial cells (NS) and 14‐dpi syncytia (S) induced in the roots of the wild‐type cultivar ‘MoneyMaker’ (A) or of transgenic plant lines NGB 6/2 (B) and NGB 5A/8 (C) containing NGB‐silencing constructs. CW, cell wall; NS, non‐syncytial cell; S, syncytial element. Arrowheads point to ribosomes. Scale bars, 1 μm.
Fig. S7 Localization of NGB (A, B) and NAB/ERabp1 (C, D) transcripts in uninfected roots of wild‐type tomato plants using in situ hybridization. Sections were hybridized with antisense probes (A, C) or with sense probes as negative controls (B, D). Green dispersed coloration (red arrows) produced by fluorescein fluorescence indicates the sites in which antisense probe hybridized to NAB/ERabp1 mRNA (C). Scale bars, 20 μm.
Fig. S8 Relative susceptibility of silenced transgenic homozygous tomato lines to Globodera rostochiensis pathotype Ro1 in pot trials. Values are expressed as a percentage of the number of females that developed on transgenic tomato roots relative to the number on control roots. The number of females on control roots was taken to be 100%.
Fig. S9 Measurements of main root lengths in transgenic tomato lines NGB and NAB/ERabp1. Seeds were grown on 0.8% agar Petri dishes and were placed in a dark growth chamber at a temperature of 18 °C for 5‐7 days to elicit seed germination. Next, germinated seeds were transferred to 0.5 × Murashige and Skoog (MS) solid medium (pH 6.2) and grown in a chamber under controlled conditions (light : dark photoperiod, 16 h : 8 h; warm : cold thermoperiod, 25 °C : 18 °C; light intensity, 54 μmol/m2/s). After 2 weeks, the main roots were measured using a dissecting microscope (Leica M5C, Wentzlar, Germany), and photographic documentation was obtained using a specialized digital camera (Leica DFC 425, Wentzlar, Germany). Results are presented as the mean values (± SE) obtained from 15 roots per line. The asterisks indicate the significance of the differences from the control as revealed by one‐way analysis of variance (ANOVA) test: *P < 0.05.
Table S1 Efficiency of tomato transformation with NGB or NAB/ERabp1 silencing constructs.
Table S2 Segregation analysis of tomato T1 seedlings during kanamycin selection. Plants were obtained following transformation with NGB or NAB/ERabp1 gene‐silencing constructs. Resistant seedlings possessed green (not purple) hypocotyls and developed true leaves and lateral roots after 1 month of culture. Results were verified using the χ 2 test and the following null hypothesis: if the resistant : susceptible ratio is 3:1, then a transgene is considered to have been incorporated at a single locus. P 0.05 = 3.84; one degree of freedom.
Table S3 Characteristics of primers used in real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) to verify NGB and NAB/ERabp1 gene expression profiles in transgenic and wild‐type tomato plants.
Table S4 Characteristics of primers used in real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) to verify expression profiles of auxin markers and pathogenesis‐related genes in transgenic tomato lines NGB 6/2 and NAB/ERabp1 3/9.
Table S5 Reaction mixture composition used in real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of NGB and NAB/ERabp1 expression.
Table S6 Reaction mixture composition used in real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of expression profiles of auxin markers and pathogenesis‐related genes in transgenic tomato lines NGB 6/2 and NAB/ERabp1 3/9.
Table S7 Real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) conditions used for analysis of NGB and NAB/ERabp1 expression profiles.
Table S8 Real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) conditions used for analysis of expression profiles of auxin markers and pathogenesis‐related genes in transgenic tomato lines NGB 6/2 and NAB/ERabp1 3/9.
Acknowledgements
This work was supported by grants from the Polish Ministry of Science and Higher Education (PBZ‐KBN‐089/P06/2003, NN302060534 and 116/N‐COST/2008/0), the Polish National Science Center (N N302 593938) and by the WULS‐PLANT HEALTH project (FP7‐286093). We thank Dr Aska Goverse (Laboratory of Nematology, Wageningen University, the Netherlands) for kindly sharing nematode cysts, Malgorzata Wasilewska‐Gomulka (Department of Botany, Warsaw University of Life Sciences, Warsaw, Poland) for ultramicrotomy assistance, Karolina Morgiewicz and Anna Mędrzycka (Department of Plant Physiology, Warsaw University of Life Sciences, Warsaw, Poland) for assistance in the generation of homozygous tomato plants and Dr Anna Przetakiewicz (Laboratory of Quarantine Organisms, Department of Phytopathology, Plant Breeding and Acclimatization Institute, Radzikow, Poland) for setting up the pot trials.
References
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. and Lipman, D.J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Anderson, D.J. and Botella, J.R. (2007) Expression analysis and subcellular localization of Arabidopsis thaliana G‐protein β‐subunit AGB1. Plant Cell Rep. 26, 1469–1480. [DOI] [PubMed] [Google Scholar]
- Assmann, S.M. (2002) Heterotrimeric and unconventional GTP binding proteins in plant cell signaling. Plant Cell, 14, 355–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann, S.M. (2005) G protein go green: a plant G protein signaling FAQ sheet. Science, 310, 71–73. [DOI] [PubMed] [Google Scholar]
- Chomczynski, P. (1993) A reagent for the single step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Bio Techniques, 15, 532–537. [PubMed] [Google Scholar]
- David, K.M. , Couch, D. and Perrot‐Rechenmann, C. (2007) Does auxin binding protein 1 control both cell division and cell expansion? Plant Signal. Behav. 2, 376–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effendi, Y. , Rietz, S. , Fischer, U. and Scherer, G.F.E. (2011) The heterozygous abp1/ABP1 insertional mutant has defects in functions requiring polar auxin transport and in regulation of early auxin‐regulated genes. Plant J. 65, 282–294. [DOI] [PubMed] [Google Scholar]
- Expósito‐Rodríguez, M. , Borges, A.A. , Borges‐Pérez, A. and Pérez, J.A. (2008) Selection of internal control genes for quantitative real‐time RT‐PCR studies during tomato development process. BMC Plant Biol. 8, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frary, A. and Earle, E.D. (1996) An examination of factors affecting the efficiency of Agrobacterium‐mediated transformation of tomato. Plant Cell Rep. 16, 235–240. [DOI] [PubMed] [Google Scholar]
- Fudali, S. , Janakowski, S. , Sobczak, M. , Griesser, M. , Grundler, F.M.W. and Golinowski, W. (2008) Two tomato α‐expansins show distinct spatial and temporal expression patterns during development of nematode‐induced syncytia. Physiol. Plant. 132, 370–383. [DOI] [PubMed] [Google Scholar]
- Fuentes, J.L. , Datta, K. , Sullivan, S.M. , Walker, A. and Maddock, J.R. (2007) In vivo functional characterization of the Saccharomyces cerevisiae 60S biogenesis GTPase Nog1. Mol. Genet. Genomics, 278, 105–123. [DOI] [PubMed] [Google Scholar]
- Gheysen, G. and Mitchum, M.G. (2009) Molecular insights in the susceptible plant response to nematode infection In: Cell Biology of Plant Nematode Parasitism. Plant Cell Monographs (Berg R.H. and Taylor C.G., eds), p. 45 New York: Springer. [Google Scholar]
- Gheysen, G. and Vanholme, B. (2007) RNAi from plant to nematodes. Trends Biotechnol. 25, 89–92. [DOI] [PubMed] [Google Scholar]
- Gheysen, G. , van der Eycken, W. , Barthels, N. , Karimi, M. and van Montagu, M. (1996) The exploitation of nematode‐responsive plant genes in novel nematode control methods. Pestic. Sci. 47, 95–101. [Google Scholar]
- Golinowski, W. , Grundler, F.M.W. and Sobczak, M. (1996) Changes in the structure of Arabidopsis thaliana during female development of the plant‐parasitic nematode Heterodera schachtii . Protoplasma, 194, 103–116. [Google Scholar]
- Goverse, A. , Engler, J. , Verhees, J. , van der Krol, S. , Helder, J. and Gheysen, G. (2000a) Cell cycle activation by plant parasitic nematodes. Plant Mol. Biol. 43, 747–761. [DOI] [PubMed] [Google Scholar]
- Goverse, A. , Overmars, H. , Engelbertink, J. , Schots, A. , Bakker, J. and Helder, J. (2000b) Both induction and morphogenesis of cyst nematode feeding cells are mediated by auxin. Mol. Plant–Microbe Interact. 13, 1121–1129. [DOI] [PubMed] [Google Scholar]
- Grunewald, W. , Karimi, M. , Wieczorek, K. , Van de Capelle, E. , Grundler, F. , Beeckman, T. , Inze, D. and Gheysen, G. (2008) A role of AtWRKY23 in feeding site establishment of plant‐parasitic nematodes. Plant Physiol. 148, 358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald, W. , Cannoot, B. , Friml, J. and Gheysen, G. (2009a) Parasitic nematodes modulate PIN‐mediated auxin transport to facilitate infection. PLoS Pathog. 5, e1000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald, W. , Van Noorden, G. , Van Listerdael, G. , Beeckman, T. and Gheysen, G. (2009b) Manipulation of auxin transport in plant roots during rhizobium symbiosis and nematode parasitism. Plant Cell, 21, 2553–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegeman, A. , Mantelin, S. , Jones, J.T. and Gheysen, G. (2012) Functional roles of effectors of plant‐parasitic nematodes. Gene, 15, 492, 19–31. [DOI] [PubMed] [Google Scholar]
- Hermsmeier, D. , Mazarei, M. and Baum, T.J. (1998) Differential display analysis of the compatible interaction between soybean and the soybean cyst nematode. Mol. Plant–Microbe Interact. 13, 309–315. [Google Scholar]
- Hesse, T. , Feldwisch, J. , Balshusemann, D. , Bauw, G. , Puype, M. , Vandekerckhove, J. , Lobler, M. , Klambt, D. , Schell, J. and Palme, K. (1989) Molecular cloning and structural analysis of a gene from Zea mays (L.) coding for a putative receptor for the plant hormone auxin. EMBO J. 8, 2453–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, G. (2006) Engineering broad root‐knot resistance in transgenic plants by RNAi silencing of a conserved and essential root‐knot nematode parasitism gene. Proc. Natl. Acad. Sci. USA, 103, 14 302–14 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, B.C. , Wang, Q. , Kifer, C.T. and Parsons, M. (2003) The NOG1 GTP‐binding protein is required for biogenesis of the 60S ribosomal subunit. J. Biol. Chem. 278, 32 204–32 211. [DOI] [PubMed] [Google Scholar]
- Jones, A.M. (1994) Auxin‐binding proteins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 45, 393–420. [Google Scholar]
- Kandoth, P.K. , Ithal, N. , Recknor, J. , Maier, T. , Nettleton, D. , Baum, T.J. and Mitchum, M.G. (2011) The soybean Rhg1 locus for resistance to the soybean cyst nematode Heterodera glycines regulates the expression of a large number of stress‐ and defense‐related genes in degeneration feeding cells. Plant Physiol. 155, 1960–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczmarek, A. , Fudali, S. , Lichocka, M. , Sobczak, M. , Kurek, W. , Janakowski, S. , Roosien, J. , Golinowski, W. , Bakker, J. , Goverse, A. and Helder, J. (2008) Expression of two functionally distinct plant endo‐β‐1,4‐glucanases is essential for the compatible interaction between potato cyst nematode and its host. Mol. Plant–Microbe Interact. 21, 791–798. [DOI] [PubMed] [Google Scholar]
- Letunic, I. , Doerks, T. and Bork, P. (2012) SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40, D302–D305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Fester, T. and Taylor, C. (2009) Transcriptomic analysis of nematode infestation In: Cell Biology of Plant Nematode Parasitism. Plant Cell Monographs (Berg R.H. and Taylor C.G., eds), pp. 189–220. New York: Springer. [Google Scholar]
- Lilley, C.J. , Davies, L.J. and Urwin, P.E. (2012) RNA interference in plant parasitic nematodes: a summary of the current status. Parasitology, 139, 630–640. [DOI] [PubMed] [Google Scholar]
- Liu, J. , Ding, P. , Sun, T. , Nitta, Y. , Dong, O. , Huang, X. , Yang, W. , Li, X. , Botella, J.R. and Zhang, Y. (2013) Heterotrimeric G proteins serve as a converging point in plant defense signaling activated by multiple receptor‐like kinases. Plant Physiol. 161, 2146–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano‐Torres, J.L. , Wilbers, R.H.P. , Gawronski, P. , Boshoven, J.C. , Finkers‐Tomczak, A. , Cordewener, J.H.G. , America, A.H.P. , Overmars, H.A. , van't Klooster, J.W. , Baranowski, L. , Sobczak, M. , Ilyas, M. , van der Hoorn, R.A.L. , Schots, A. , de Wit, P.J.G.M. , Bakker, J. , Goverse, A. and Smant, G. (2013) Dual disease resistance mediated by the immune receptor Cf‐2 in tomato requires a common virulence target of a fungus and a nematode. Proc. Natl. Acad. Sci. USA, 109, 10 119–10 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler‐Bauer, A. , Lu, S. , Anderson, J.B. , Chitsaz, F. , Derbyshire, M.K. , DeWeese‐Scott, C. , Fong, J.H. , Geer, L.Y. , Geer, R.C. , Gonzales, N.R. , Gwadz, M. , Hurwitz, D.I. , Jackson, J.D. , Ke, Z. , Lanczycki, C.J. , Lu, F. , Marchler, G.H. , Mullokandor, M. , Omelchenko, M.V. , Robertson, C.L. , Song, J.S. , Thanki, N. , Yamashita, R.A. , Zhang, D. , Zhang, N. , Zheng, C. and Bryant, S.H. (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–D229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montillet, J.L. , Leonhardt, N. , Mondy, S. , Tranchimand, S. , Rumeau, D. , Boudsocq, M. , Garcia, A.V. , Douki, T. , Bigeard, J. , Laurière, C. , Cheralier, A. , Castresana, C. and Hirt, H. (2013) An abscisic acid‐independent oxylipin pathway controls stomatal closure and immune defense in Arabidopsis. PLoS Biol. 11, e1001513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T. and Skoog, F.A. (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Palme, K. , Feldwisch, J. , Hesse, T. , Bauw, G. , Puype, M. , Vandekerchkhove, J. and Schell, J. (1990) Auxin binding proteins from maize coleoptiles: purification and molecular characterization. Symp. Soc. Exp. Biol. 44, 299–313. [PubMed] [Google Scholar]
- Perrot‐Rechenmann, C. (2010) Cellular responses to auxin: division versus expansion. Cold Spring Harb. Perspect. Biol. 2, 63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka, J. and Nelson, T. (2007) Arabidopsis nucleolin affects plant development and patterning. Plant Physiol. 144, 173–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl, M.W. , Horgan, G.W. and Dempfle, L. (2002) Relative Expression Software Tool (REST©) for group wise comparison and statistical analysis of relative expression results in real‐time PCR. Nucleic Acids Res. 30, E36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, J. , Wen, L. , Gao, X. , Jin, C. , Xue, Y. and Yao, X. (2009) DOG 1.0: illustrator of protein domain structures. Cell Res. 19, 271–273. [DOI] [PubMed] [Google Scholar]
- Rice, S.L. , Leadbeater, B.S.C. and Stone, A.R. (1985) Changes in cell structure in roots of resistant potatoes parasitized by potato cyst nematodes. I. Potatoes with resistance gene H1 derived from Solanum tuberosum ssp. andigena . Physiol. Plant Pathol. 27, 219–234. [Google Scholar]
- Rosado, A. , Sohn, E.J. , Drakakaki, G. , Pan, S. , Swidergal, A. , Xiong, Y. , Kang, B.H. , Bressan, R.A. and Raikhel, N.V. (2010) Auxin‐mediated ribosomal biogenesis regulates vacuolar trafficking in Arabidopsis. Plant Cell, 22, 143–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist, C.J.A. , Cerutti, L. , Hulo, N. , Gattiker, A. , Falquet, L. , Pagni, M. , Bairoch, A. and Bucher, P. (2002) PROSITE: a documented database using patterns and profiles as motif descriptors. Brief. Bioinformatics, 3, 265–274. [DOI] [PubMed] [Google Scholar]
- Sindhu, A.S. , Maier, T.R. , Mitchum, M.G. , Hussey, R.S. , Davis, E.L. and Baum, T.J. (2009) Effective and specific in planta RNAi in cyst nematodes: expression interference of four parasitism genes reduces parasitic success. J. Exp. Bot. 60, 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczak, M. , Avrova, A. , Jupowicz, J. , Phillips, M.S. , Ernst, K. and Kumar, A. (2005) Characterization of susceptibility and resistance responses to potato cyst nematode (Globodera spp.) infection of tomato lines in the absence and presence of the broad‐spectrum nematode resistance Hero gene. Mol. Plant–Microbe Interact. 18, 158–168. [DOI] [PubMed] [Google Scholar]
- Suwastika, I.N. , Denawa, M. , Hata, A. , Ohniwa, R.L. , Takeyasu, K. and Shiina, T. (2008) Localization of Obg‐Hflx and TrmE‐Era super family small GTPases in various organelles in plant cells In: Photosynthesis. Energy from the Sun: 14th International Congress on Photosynthesis (Allen J.F., Gantt E., Golbeck J.H. and Osmond B., eds), pp. 1137–1140. Yokohama: Springer. [Google Scholar]
- Swiecicka, M. , Filipecki, M. , Lont, D. , Van Vliet, J. , Qin, L. , Goverse, A. , Bakker, J. and Helder, J. (2009) Dynamics in the tomato root transcriptome on infection with the potato cyst nematode Globodera rostochiensis . Mol. Plant Pathol. 10, 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomczak, A. , Koropacka, K. , Smant, G. , Goverse, A. and Bakker, E. (2009) Resistant plant responses In: Plant Cell Monographs (Berg R.H. and Taylor C.G., eds), pp. 83–113. New York/Heidelberg: Springer. [Google Scholar]
- Torres, M.A. , Morales, J. , Sánchez‐Rodríguez, C. , Molina, A. and Dangl, J.L. (2013) Functional interplay between Arabidopsis NADPH oxidases and heterotrimeric G protein. Mol. Plant–Microbe Interact. 26, 686–694. [DOI] [PubMed] [Google Scholar]
- Tromas, A. , Braun, N. , Muller, P. , Khodus, T. , Paponov, I.A. , Palme, K. , Ljung, K. , Lee, J.‐Y. , Benfey, P. , Murray, J.A.H. , Scheres, B. and Perrot‐Rechenmann, C. (2009) The Auxin Binding Protein 1 is required for differential auxin responses mediating root growth. PLoS ONE, 4, e6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudgill, D.L. (1991) Resistance to and tolerance of plant parasitic nematodes in plants. Annu. Rev. Phytopathol. 29, 167–193. [Google Scholar]
- Urwin, P.E. , Troth, K.M. , Zubko, E.I. and Atkinson, H.J. (2001) Effective transgenic resistance to Globodera pallida in potato field trials. Mol. Breed. 8, 95–101. [Google Scholar]
- Vanholme, B. , De Meutter, J. , Tytgat, T. , Van Montagu, M. , Coomans, A. and Gheysen, G. (2004) Secretions of plant‐parasitic nematodes: a molecular update. Gene, 332, 13–27. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Replogle, A. , Davis, E.L. and Mitchum, M.G. (2007) The tobacco Cel7 gene promoter is auxin‐responsive and locally induced in nematode feeding sites of heterologous plants. Mol. Plant Pathol. 8, 423–436. [DOI] [PubMed] [Google Scholar]
- Wieczorek, K. , Hoffman, J. , Bloch, A. , Szakasist, D. , Bohlman, H. and Grundler, F.M.W. (2008) Arabidopsis endo‐1,4‐glucanases are involved in the formation of root syncytia induced by Heterodera schachtii . Plant J. 53, 336–351. [DOI] [PubMed] [Google Scholar]
- Wroblewski, T. , Tomczak, A. and Michelmore, R.W. (2005) Optimization of Agrobacterium‐mediated transient assays of gene expression in lettuce, tomato and Arabidopsis . Plant Biotechnol. J. 3, 259–273. [DOI] [PubMed] [Google Scholar]
- Wroblewski, T. , Piskurewicz, U. , Tomczak, A. , Ochoa, O. and Michelmore, R.W. (2007) Silencing of the major family of NBS‐LRR‐encoding genes in lettuce results in the loss of multiple resistance specificities. Plant J. 51, 803–818. [DOI] [PubMed] [Google Scholar]
- Yadav, B.C. , Veluthambi, K. and Subramaniam, K. (2006) Host‐generated double stranded RNA induces RNAi in plant‐parasitic nematodes and protects the host from infection. Mol. Biochem. Parasitol. 148, 219–222. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Schematic diagram of predicted protein sequences encoded by NAB/ERabp1 (A) and NGB (B). The NAB/ERabp1 protein sequence possesses a highly conserved KDEL motif, a signal of endoplasmic reticulum localization, near the C‐terminus (A). The NGB protein sequence contains a NOG domain in its central part. This is characteristic of low‐molecular‐weight GTP‐binding proteins belonging to the Ras superfamily. NOGCT is a NOG C‐terminal domain. An EF‐HAND‐1 domain (EF‐hand calcium‐binding domain) involved in Ca2+ binding is located near the protein's C‐terminus (B). Both domains are activators of several proteins involved in cell‐to‐cell signal transduction.
Fig. S2 Architecture of silencing constructs in pDraCULA (Wroblewski et al., 2007). The pDraCULA vector contains the inverted repeats of a reporter gene, uidA, in order to determine the correct function of the silencing constructs. Transcription of the hairpin structure (dsRNA) is controlled by the CaMV35S promoter and a polyadenylation signal from Agrobacterium tumefaciens octopine synthase (OCS). The selection marker, nptII, conferring kanamycin resistance, is controlled by the A. tumefaciens nopaline synthase (NOS) promoter and polyadenylation signal present in the T‐DNA region. The lengths of the target gene silencing fragments were 138 bp (NGB) and 303 bp (NAB/ERabp1).
Fig. S3 Transient expression of silencing constructs in Nicotiana benthamiana leaves. Tobacco leaves infiltrated with Agrobacterium tumefaciens C58C1 strain containing the reporter construct 35S::uidA (left side of each leaf) or the reporter construct 35S::uidA (right side of each leaf) and a gene‐silencing construct pDraCULA::NAB/ERabp1SfiI (A) or pDraCULA::NGBSfiI (B). Scale bars, 1.5 cm. The silencing activity of NGB and NAB/ERabp1 gene constructs in pDraCULA was checked in N. benthamiana leaves in a transient transformation assay (Wroblewski et al., 2005). The inclusion of inversely repeated fragments of the reporter gene uidA allows the verification of correct vector functioning before stable plant transformation, as the target genes were driven by the same constitutive CaMV35S promoter. The transient assay was carried out by co‐infiltration of N. benthamiana leaves with A. tumefaciens strains harbouring the silencing constructs and monitoring of uidA gene activity.
Fig. S4 Polymerase chain reaction (PCR) results confirming the presence of transgenes in homozygous T1 tomato plants. PCR was carried out using gene‐specific primer pairs for uidA and nptII to confirm the presence of the uidA reporter gene in sense (A) and antisense (B) orientations, and the nptII gene (C) in the genomes of selected transgenic tomato plants transformed with NGB or NAB/ERabp1 gene‐silencing constructs. Fragments of the expected length (300 bp for uidA and 700 bp for nptII) were amplified and PCR products were analysed using gel electrophoresis. Abbreviations: M, DNA marker (SMO331, MBI Fermentas); N, genomic DNA from wild‐type tomato plant (negative control); P1, plasmid DNA 35S::NGB/pDraCULA (positive control); P2, plasmid DNA 35S::NAB/ERabp1/pDraCULA (positive control); 1, 2, genomic DNA of transgenic tomato plants containing the NGB‐silencing construct (lines: 1, NGB 6/2; 2, NGB 5A/8); 3–5, genomic DNA of transgenic tomato plants with the NAB/ERabp1‐silencing construct (lines: 3, NAB/ERabp1 1/10; 4, NAB/ERabp1 3/9; 5, NAB/ERabp1 8/7). The primer sequences used for the amplification of uidA and nptII genes were as follows: nptIIF, 5′‐GAGGCTATTCGGCTATGACTG‐3′; nptIIR, 5′‐ATCGGGAGCGGCGATACCGTA‐3′; uidAF, 5′‐TACACCACGCCGAACACCTG‐3′; uidAR, 5′‐AAACAAATGCCTAAAGAGAG‐3′. Primer annealing temperature, 55 °C; elongation time, 30 s for uidA and 60 s for nptII.
Fig. S5 Expression levels of NGB and NAB/ERabp1 in leaves of homozygous T2 tomato plants carrying gene‐silencing constructs. The silencing effect of the constructs on target gene expression in homozygous T2 tomato lines was determined by real‐time reverse transcription‐polymerase chain reaction (RT‐PCR); expression levels of NGB and NAB/ERabp1 were quantified with reference to the expression of 18S rRNA. The stability of gene silencing was confirmed in transgenic lines selected for further examination. In homozygous T2 plants, we observed a reduction in NGB expression of 76% in NGB lines 5A/8 and 6/2, a reduction of 37% and in NAB/ERabp1 lines 3/9, 8/7 and 1/10, a reduction in NAB/ERabp1 expression of 78%, 88% and 73%, respectively. Relative expression levels are shown as fold changes in comparison with the expression of a given mRNA in a control sample that was arbitrarily assigned a value of unity. The results presented are means (± SE) obtained from three independent experiments.
Fig. S6 Comparison of cytoplasm ultrastructure and ribosomal abundance between neighbouring non‐syncytial cells (NS) and 14‐dpi syncytia (S) induced in the roots of the wild‐type cultivar ‘MoneyMaker’ (A) or of transgenic plant lines NGB 6/2 (B) and NGB 5A/8 (C) containing NGB‐silencing constructs. CW, cell wall; NS, non‐syncytial cell; S, syncytial element. Arrowheads point to ribosomes. Scale bars, 1 μm.
Fig. S7 Localization of NGB (A, B) and NAB/ERabp1 (C, D) transcripts in uninfected roots of wild‐type tomato plants using in situ hybridization. Sections were hybridized with antisense probes (A, C) or with sense probes as negative controls (B, D). Green dispersed coloration (red arrows) produced by fluorescein fluorescence indicates the sites in which antisense probe hybridized to NAB/ERabp1 mRNA (C). Scale bars, 20 μm.
Fig. S8 Relative susceptibility of silenced transgenic homozygous tomato lines to Globodera rostochiensis pathotype Ro1 in pot trials. Values are expressed as a percentage of the number of females that developed on transgenic tomato roots relative to the number on control roots. The number of females on control roots was taken to be 100%.
Fig. S9 Measurements of main root lengths in transgenic tomato lines NGB and NAB/ERabp1. Seeds were grown on 0.8% agar Petri dishes and were placed in a dark growth chamber at a temperature of 18 °C for 5‐7 days to elicit seed germination. Next, germinated seeds were transferred to 0.5 × Murashige and Skoog (MS) solid medium (pH 6.2) and grown in a chamber under controlled conditions (light : dark photoperiod, 16 h : 8 h; warm : cold thermoperiod, 25 °C : 18 °C; light intensity, 54 μmol/m2/s). After 2 weeks, the main roots were measured using a dissecting microscope (Leica M5C, Wentzlar, Germany), and photographic documentation was obtained using a specialized digital camera (Leica DFC 425, Wentzlar, Germany). Results are presented as the mean values (± SE) obtained from 15 roots per line. The asterisks indicate the significance of the differences from the control as revealed by one‐way analysis of variance (ANOVA) test: *P < 0.05.
Table S1 Efficiency of tomato transformation with NGB or NAB/ERabp1 silencing constructs.
Table S2 Segregation analysis of tomato T1 seedlings during kanamycin selection. Plants were obtained following transformation with NGB or NAB/ERabp1 gene‐silencing constructs. Resistant seedlings possessed green (not purple) hypocotyls and developed true leaves and lateral roots after 1 month of culture. Results were verified using the χ 2 test and the following null hypothesis: if the resistant : susceptible ratio is 3:1, then a transgene is considered to have been incorporated at a single locus. P 0.05 = 3.84; one degree of freedom.
Table S3 Characteristics of primers used in real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) to verify NGB and NAB/ERabp1 gene expression profiles in transgenic and wild‐type tomato plants.
Table S4 Characteristics of primers used in real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) to verify expression profiles of auxin markers and pathogenesis‐related genes in transgenic tomato lines NGB 6/2 and NAB/ERabp1 3/9.
Table S5 Reaction mixture composition used in real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of NGB and NAB/ERabp1 expression.
Table S6 Reaction mixture composition used in real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of expression profiles of auxin markers and pathogenesis‐related genes in transgenic tomato lines NGB 6/2 and NAB/ERabp1 3/9.
Table S7 Real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) conditions used for analysis of NGB and NAB/ERabp1 expression profiles.
Table S8 Real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) conditions used for analysis of expression profiles of auxin markers and pathogenesis‐related genes in transgenic tomato lines NGB 6/2 and NAB/ERabp1 3/9.


