Figure 1.
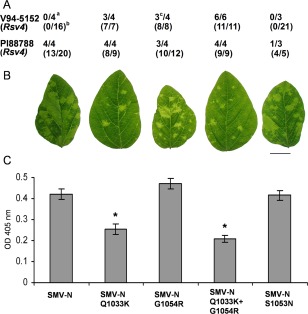
(A) Virulence of SMV‐N and its derivative P3 mutants on Rsv4‐genotype soybeans inoculated biolistically with an infectious cDNA clonea or mechanically with infectious sap derived from biolistically inoculated Essex (rsv4)b; the number of plants systemically infected/number of plants inoculated is shown (Khatabi et al., 2012; Wang et al., 2015). cInfection of V94‐5152 (Rsv4) with SMV‐NG1054R was associated with a sequence polymorphism in P3 of progeny viruses at position 3265, in which both GTA and GCA codons, encoding valine and alanine, respectively, were present at polyprotein position 1045 (Khatabi et al., 2012). (B) Systemic symptoms on central leaflets from trifoliate leaves of soybean cv. Essex (rsv4) inoculated mechanically on unifoliate leaves with progeny viruses derived from cDNA clones. Plants were maintained in a growth chamber at 22°C until being photographed at 21 days post‐inoculation (dpi). Scale bar, 2 cm. (C) Comparison of the accumulation level of viruses relative to SMV‐N in systemically infected trifoliate leaves of soybean cv. Essex (rsv4) by enzyme‐linked immunosorbent assay (ELISA). Plants were inoculated and maintained as in (B) until central leaflets from four fully developed trifoliate leaves of the inoculated plants were harvested at 21 dpi and analysed. Each bar represents the mean value of virion accumulation from two independent experiments, each with four replicate plants, with the standard errors indicated. Significant differences between the mean values for SMV‐N and each of its derivative P3 mutants were determined using Kruskal–Wallis test [non‐parametric statistical method from Data Processing System software (Tang and Zhang, 2013)] and are indicated by asterisks (P < 0.05). OD, optical density; SMV, Soybean mosaic virus.
