Summary
The ribosome‐inactivating protein BE27 from sugar beet (Beta vulgaris L.) leaves is an apoplastic protein induced by signalling compounds, such as hydrogen peroxide and salicylic acid, which has been reported to be involved in defence against viruses. Here, we report that, at a concentration much lower than that present in the apoplast, BE27 displays antifungal activity against the green mould Penicillium digitatum, a necrotrophic fungus that colonizes wounds and grows in the inter‐ and intracellular spaces of the tissues of several edible plants. BE27 is able to enter into the cytosol and kill fungal cells, thus arresting the growth of the fungus. The mechanism of action seems to involve ribosomal RNA (rRNA) N‐glycosylase activity on the sarcin–ricin loop of the major rRNA which inactivates irreversibly the fungal ribosomes, thus inhibiting protein synthesis. We compared the C‐terminus of the BE27 structure with antifungal plant defensins and hypothesize that a structural motif composed of an α‐helix and a β‐hairpin, similar to the γ‐core motif of defensins, might contribute to the specific interaction with the fungal plasma membranes, allowing the protein to enter into the cell.
Keywords: apoplast, defensin, green mould, plant defence, pokeweed antiviral protein (PAP), polynucleotide:adenosine glycosylase, rRNA N‐glycosylase
Introduction
Plants have developed protein‐based defences against infections by fungi and other pathogens. Some of these proteins are common and have been classified into 17 families of pathogenesis‐related (PR) proteins, and others occur specifically in some plant species (van Loon et al., 2006). Most are induced by signalling compounds, such as salicylic acid, jasmonic acid, ethylene or hydrogen peroxide, and display antimicrobial activities in vitro through different enzymatic activities (van Loon et al., 2006).
The plant apoplast plays a crucial role in plant defence as it contains proteins that trigger the plant defence response and inducible enzymes that are toxic to fungi, such as proteases, chitinases, glucanases, reactive oxygen species (ROS)‐generating enzymes and defensins (Doehlemann and Hemetsberger, 2013).
The apoplast of sugar beet (Beta vulgaris L.) leaves also contains the ribosome‐inactivating proteins (RIPs) BE27 and BE29 which represent different levels of glycosylation of the same protein (Iglesias et al., 2005). These proteins are synthesized in response to virus infection (Girbes et al., 1996) and are expressed in both infected (hypersensitive response) and non‐infected (systemic acquired resistance, SAR) leaves (Iglesias et al., 2005). Furthermore, the signalling compounds hydrogen peroxide and salicylic acid induce the expression of both proteins (Iglesias et al., 2005, 2008). The hypothesis that BE27 is involved in plant defence against viruses is partially supported, although in non‐physiological conditions, by the fact that the external application of BE27 to sugar beet leaves prevents infection by Artichoke mottled crinkle virus (AMCV) and because this protein displays polynucleotide:adenosine glycosylase activity against Tobacco mosaic virus (TMV) genomic RNA (gRNA) (Iglesias et al., 2005).
RIPs are protein synthesis inhibitors that irreversibly inactivate mammalian ribosomes (Girbes et al., 2004; Puri et al., 2012). RIPs can also inhibit protein synthesis in other animals and yeasts (Girbes et al., 2004), and some can inactivate plant and bacterial ribosomes, although usually these ribosomes are insensitive to the majority of RIPs (Girbes and Ferreras, 1998). These proteins are 28S ribosomal RNA (rRNA) N‐glycosylases (EC 3.2.2.22) which cleave the N‐glycosidic bond between adenine at position 4324 and its ribose in the 60S subunit of rat ribosomes. This adenine is located in the sarcin–ricin loop (SRL), which is crucial for anchoring the EFG or EF‐2 elongation factors on the ribosome during messenger RNA–transfer RNA (mRNA–tRNA) translocation in prokaryotes and eukaryotes, respectively (Girbes et al., 2004; Puri et al., 2012). In addition, some RIPs release adenines from viral gRNAs (Barbieri et al., 1997; Iglesias et al., 2005) and herring sperm DNA (Barbieri et al., 1997; Iglesias et al., 2015). Thus, Barbieri et al. (1997) have proposed the name polynucleotide:adenosine glycosylases for these proteins and have suggested that extensive deadenylation of viral RNA and DNA might be responsible for the antiviral action of RIPs. From a structural point of view, RIPs have been classified into two types: type 1 [e.g. saporin, pokeweed antiviral protein (PAP), BE27 and ME] consisting of single‐chain proteins, and type 2 (e.g. ricin, abrin and ebulin) consisting of an A (active) chain with RIP properties covalently linked to a B (binding) chain with lectin properties. The latter RIPs can enter cells more easily because the B chain allows binding to sugar‐containing cell surface receptors and, for this reason, type 2 RIPs can be potent toxins, e.g. ricin, abrin or volkensin (Citores et al., 2013; Ferreras et al., 2011a). Type 1 RIPs from monocots lack the leader peptide (Di Maro et al., 2014) and, for this reason, these proteins are synthesized and localized in the cytosol (Chaudhry et al., 1994; Lanzanova et al., 2011). By contrast, type 1 RIPs from dicots and type 2 RIPs contain a leader peptide (Di Maro et al., 2014), thus being synthesized in the endoplasmic reticulum. Several studies have indicated that type 1 RIPs from dicots are localized exclusively (Di Cola et al., 1999; Iglesias et al., 2005; Park et al., 2004; Yoshinari et al., 1996) or mainly (Carzaniga et al., 1994; Parente et al., 2008; Yoshinari et al., 1997) in the apoplast, whereas type 2 RIPs are localized in protein storage vacuoles (Lord and Spooner, 2011).
RIPs have been extensively researched because of their ability to inhibit protein synthesis and their utility as antiviral (Fang et al., 2011) and antitumour (Pan et al., 2014) agents. The most promising applications of RIPs in experimental medicine, especially in anticancer therapy, are related to their use as components of immunotoxins, conjugates or recombinant chimeras in which they are linked to tumour‐targeting ligands or antibodies which mediate their binding and internalization by malignant cells (Becker and Benhar, 2012; Citores et al., 2013; Ferreras et al., 2011a; Polito et al., 2011). However, some RIPs behave as defence proteins as they are induced by salt stress (Rippmann et al., 1997), insect elicitors (Engelberth et al., 2012), mechanical wounding (Engelberth et al., 2012; Song et al., 2000), pathogenic fungus (Tan et al., 2013) and temperature and ultraviolet light (Qin et al., 2009). In addition, some are induced by signalling compounds, such as jasmonic acid (Chaudhry et al., 1994; Song et al., 2000), salicylic acid (Qin et al., 2009) or abscisic acid (Song et al., 2000). Furthermore, several studies have indicated that RIPs protect plants from viral (Park et al., 2004; Tumer et al., 1997) and fungal (Park et al., 2002) infection, and transgenic plants bearing RIP genes have been constructed that are resistant to viruses, fungi and insects (Corrado et al., 2005; Dowd et al., 2012; Huang et al., 2008; Qian et al., 2014).
Recently, we have tested other activities of BE27 that could play a defensive role against pathogens (Iglesias et al., 2015), finding that BE27 displays rRNA N‐glycosylase activity against yeast and Agrobacterium tumefaciens ribosomes, DNA polynucleotide:adenosine glycosylase activity against herring sperm DNA, and magnesium‐dependent endonuclease activity against the supercoiled plasmid PUC19 (nicking activity). In addition, BE27 possesses superoxide dismutase (SOD) activity, thus being able to produce the signal compound hydrogen peroxide. BE27 is also toxic to COLO 320 cells, inducing apoptosis in these cells. The combined effect of these biological activities could result in a broad action against several types of pathogen, such as viruses (polynucleotide:adenosine glycosylase activity), bacteria (rRNA N‐glycosylase and nicking activities), fungi (rRNA N‐glycosylase activity) and insects (via the induction of gut cell apoptosis) (Iglesias et al., 2015).
In order to gain an insight into the protective properties of BE27 against pathogenic fungi, we have carried out experiments to test whether this protein is able to enter the fungal cytoplasm and arrest the growth of the fungus Penicillium digitatum, which infects a broad spectrum of edible plants, such as kiwifruit, sugarbeet, apple, plum, spinach and, mainly, citrus (http://www.plantwise.org/KnowledgeBank/Datasheet.aspx?dsid=39570) (Frisvad and Samson, 2004). At concentrations lower than those found in the apoplast, BE27 was able to enter the cytosol and kill fungal cells, thus arresting the growth of the fungus. The mechanism of action seemed to involve rRNA N‐glycosylase activity on the SRL of the large‐subunit rRNA which inactivates irreversibly the fungal ribosomes, thus inhibiting protein synthesis. In addition, we found that, close to its C‐terminus, BE27 contains a structural motif composed of an α‐helix and a β‐hairpin similar to that present in antifungal defensins. Such a β‐hairpin has been reported to be responsible for the interaction of defensins with the fungal plasma membrane, allowing these apoplastic proteins to enter into fungal cells (Lacerda et al., 1977; Sagaram et al., 2013).
Results
Effect of BE27 on the growth of P. digitatum
To characterize the antifungal properties of BE27, we carried out experiments to test the effect of this protein on the growth of the fungus P. digitatum. As illustrated in Fig. 1, BE27 showed a strong growth‐inhibiting effect on P. digitatum and reduced fungal growth in a concentration‐dependent manner. Thus, 8, 4 and 0.6 μg/mL of BE27 resulted in 70%, 57% and 24% growth inhibition, respectively, after 78 h of incubation with conidia as starting material. In addition, 8 μg/mL of BE27 added to mycelia growing for 28 h inhibited fungal growth by 84%, suggesting that BE27 has a stronger effect on mycelial growth than on conidial germination. These concentrations are lower than those reached by BE27 and BE29 in the apoplast in B. vulgaris leaves. To estimate the concentration of BE27 and BE29 present in the apoplast, we studied the translation inhibitory activity of B. vulgaris crude extracts from 6‐week‐old plants untreated or treated with either hydrogen peroxide or salicylic acid, and 7‐month‐old plants collected in a crop field (Table 1). These data must be evaluated with care as they may be overestimated or underestimated because of the presence of other active substances in the crude extracts. The crude extract obtained from 7‐month‐old plants was the least active in inhibiting protein synthesis in rabbit reticulocyte lysate. Such an extract contained 24 781 units of BE27 and BE29 per gram of tissue. From this preparation, we obtained 4.6 μg of BE27 and 7.3 μg of a mix containing approximately equal amounts of BE27 and BE29, for a total of 12 395 units/g of tissue, thus obtaining a purification yield of 50%. Considering 60 μL per gram of fresh weight as the volume of apoplastic fluid (Lopez‐Millan et al., 2000; Winter et al., 1994), the concentration of BE27/29 must be at least 200 μg/mL, much higher than that required to stop the growth of P. digitatum. The concentration was even higher in 6‐week‐old plants grown in a chamber, and increased two‐ and seven‐fold in hydrogen peroxide‐ and salicylic acid‐treated plants, respectively (Table 1). It has been shown previously that beetin expression is greatly enhanced on treatment with the signalling compounds hydrogen peroxide and salicylic acid (Iglesias et al., 2005). Thus, the apoplast, which is the first cellular compartment to be invaded in the infection process (Doehlemann and Hemetsberger, 2013), contains sufficient BE27 to arrest fungal growth.
Figure 1.
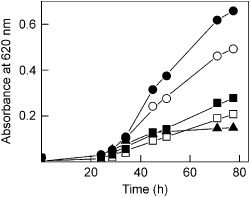
Antifungal activity of BE27 against Penicillium digitatum. Antifungal activity of BE27 against P. digitatum was measured in a microtitre plate bioassay. Conidia of P. digitatum were grown in potato dextrose broth (PDB) medium in the presence of different concentrations of BE27. Fungal growth was measured as the increase in absorbance at 620 nm. The curves represent buffer control (filled circles), 0.6 μg/mL BE27 (open circles), 4 μg/mL BE27 (filled squares) and 8 μg/mL BE27 (open squares). Conidia of P. digitatum were grown in PDB for 28 h before exposure to 8 μg/mL BE27 (filled triangles).
Table 1.
Quantification of BE27 and BE29 activity in crude extracts from Beta vulgaris leaves. Crude extracts were obtained as indicated in the Experimental procedures section. The values are referred to 1 g of fresh tissue
| Crops | Protein (μg/g of tissue) | IC50 a (ng/mL) | Specific activityb (units/mg of protein) | Total activityb (units/g of tissue) | Yieldc (%) |
|---|---|---|---|---|---|
| Control | 18500 | 310 | 3226 | 59677 | — |
| Hydrogen peroxide‐treated | 22400 | 211 | 4739 | 106161 | — |
| Salicylic acid‐treated | 18100 | 39.8 | 25125 | 454773 | — |
| Crop field | 22080 | 891 | 1122 | 24781 | (100) |
| BE27 | 4.6 | 0.96 | 1041666 | 4791 | 50.0 |
| BE27/29 | 7.3 | 0.96 | 1041666 | 7604 | — |
IC50 is the concentration giving 50% inhibition of protein synthesis in a rabbit reticulocyte lysate system.
One unit of activity is defined as the amount of protein necessary to inhibit protein synthesis by 50% in 1 mL of rabbit reticulocyte lysate reaction mixture.
The yield is referred to the purification of BE27 and BE27/29 from 300 g of leaves from a crop field.
Microscopic analysis and BE27 uptake by P. digitatum
The effect of BE27 on the mycelial growth of P. digitatum was also visualized microscopically (Fig. 2). Corresponding to growth inhibition, microscopy studies revealed modifications of hyphal morphology after exposure to BE27. Compared with the untreated control, which presented regular and homogeneous hyphae, hyper‐branching and aborted hyphal branches could be observed in the cultures treated with BE27 (Fig. 2a, left panels). Moreover, chitin staining with calcofluor white (CFW) revealed that the treatment with BE27 inhibited hyphal elongation, rendering cells significantly shorter than control cells (Fig. 2a, right panels).
Figure 2.
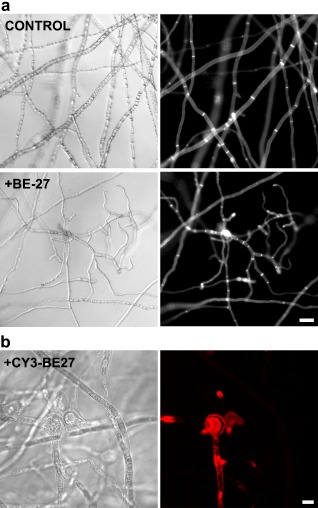
(a) Morphological changes in Penicillium digitatum mycelium exposed to BE27. Penicillium digitatum mycelium was grown in the absence (control) or presence of 5 μg/mL of BE27. After 24 h of incubation, samples were stained with calcofluor white (CFW) and visualized under the microscope. Panels represent bright‐field images (left) and fluorescence images indicative of CFW staining (right) for the same fields. Bar, 20 μm. (b) Interaction between BE27 and P. digitatum. To visualize the interaction of BE27 with fungal structures, conidia of P. digitatum were grown in vitro in potato dextrose broth (PDB) medium for 24 h before exposure to 5 μg/mL of CY3‐BE27 for 24 h. After washing, the samples were visualized with a confocal laser microscope. The red channel image (right panel) and the corresponding interference contrast image (left panel) are shown. Bar, 5 μm.
It has been reported that Trichosanthes kirilowii Maxim plant cell cultures secrete three bifunctional plant enzymes with both rRNA N‐glycosylase and chitinase activities (Shih et al., 1997). Moreover, sugar beet leaves contain chitinases that can be induced by pathogen infection (Berglund et al., 1995). To exclude the possibility that chitinase activity, either by contamination or as a consequence of an intrinsic activity of BE27, may be responsible for fungal growth inhibition, we performed experiments to assess whether our BE27 samples displayed such activity. We also tested samples with known chitinase activity, such as crude extracts from non‐induced and signalling compound‐induced sugar beet leaves, as well as the chitinase from Streptomyces griseus. The results indicated that no such activity was present in BE27 samples in standard conditions, in which the other mentioned samples gave values corresponding to the presence of chitinase activity as expected (data not shown).
Some antifungal apoplastic proteins, such as chitinases or glucanases, act outside the fungus cells (Doehlemann and Hemetsberger, 2013); however, in order to depurinate fungal ribosomes, BE27 must enter into the cytosol. In order to study the interaction of BE27 with fungal cells, the location of fluorescent‐labelled BE27 in treated hyphae was monitored by confocal microscopy. In these conditions, cyanine 3 (Cy3)‐labelled BE27 retained full antifungal activity (data not shown). On incubation with 5 μg/mL of BE27 for 24 h, BE27 accumulated in the cytoplasmic space of terminal branches of P. digitatum hyphae (Fig. 2b).
rRNA glycosylase activity of BE27 on the P. digitatum ribosomes
Because the rRNA N‐glycosylase activity of BE27 may play an antifungal role in sugar beet, we assayed the effect of BE27 on ribosomes from P. digitatum. As shown in Fig. 3, BE27 displayed rRNA N‐glycosylase activity on these ribosomes, as indicated by the release of the diagnostic fragment (Iglesias et al., 2015) on treatment with acid aniline. For comparative purposes, Fig. 3 also shows the rRNA N‐glycosylase assay on yeast. The released fragments displayed sizes around 380 and 350 nucleotides for yeast and P. digitatum, respectively, in accordance with that expected for SRL deglycosylation (368 and 359 nucleotides for yeast and Penicillium, respectively; Fig 3).Therefore, this RIP might enter into the fungal cells and inactivate their ribosomes, avoiding the propagation of the pathogen, as has been reported for ME, a type 1 RIP from Mirabilis expansa (Ruiz & Pav.) Standl. (Park et al., 2002).
Figure 3.
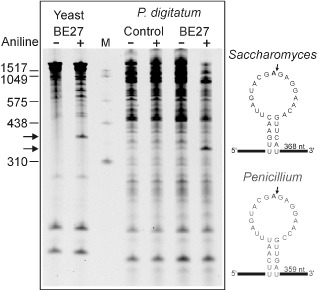
rRNA N‐glycosylase activity of BE27 on yeast and Penicillium digitatum ribosomes. Left: rRNA N‐glycosylase activity was assayed as indicated in Experimental procedures. Each lane contained 3 μg of RNA isolated from either untreated (control) or BE27‐treated ribosomes from yeast or P. digitatum. The arrows indicate the RNA fragments released as a consequence of ribosome‐inactivating protein (RIP) action on acid aniline treatment (+). Numbers indicate the size of the standards (M) in nucleotides. Right: sarcin–ricin loop of the large rRNA from yeast and Penicillium. The sequences from Saccharomyces cerevisiae (accession number J01355) and Penicillium solitum (JN642222) were downloaded from the National Center for Biotechnology Information (NCBI) sequence database (http://www.ncbi.nlm.nih.gov/nucleotide/). The large rRNA 3′ end from Penicillium was determined by alignment with the large rRNA from Saccharomyces. The adenine released by RIP action (bold type), the site of splitting by the acid aniline (arrow) and the size of the generated fragment are also indicated.
Enzymatic activity of BE27 in P. digitatum cultures
BE27, at concentrations of 4 μg/mL, showed a strong growth inhibitory effect on P. digitatum cultures in liquid medium (Figs 1 and 4a). The RNAs from these cultures were isolated and analysed to detect the presence of the RNA fragment diagnostic of RIP rRNA N‐glycosylase (Fig. 4b). The diagnostic fragment was absent in the RNA from control cultures and present in that from cultures grown in the presence of BE27. This indicates that BE27, at a concentration of 4 μg/mL, is able to enter into fungal cells in a manner that allows it to reach the cytosolic ribosomes.
Figure 4.
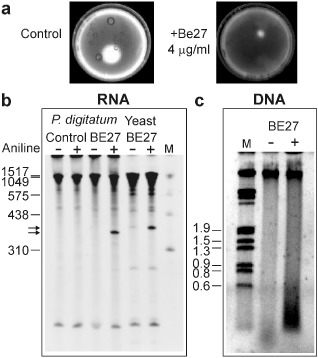
Antifungal and rRNA N‐glycosylase activity of BE27 against Penicillium digitatum. (a) Penicillium digitatum was grown in potato dextrose broth (PDB) in the absence (control) or presence of 4 μg/mL BE27 for 5 days. The mycelium was extensively washed with sterile water and harvested to extract the RNA and DNA. Representative photographs of two plates are shown. (b) rRNA N‐glycosylase activity was assayed as indicated in Experimental procedures. Each lane contained 3 μg of RNA isolated from either untreated (control) or BE27‐treated cultures from P. digitatum. For comparative purposes, yeast RNA depurinated by BE27 is also included in the assay. The arrows indicate the RNA fragments released as a consequence of ribosome‐inactivating protein (RIP) action on acid aniline treatment (+). Numbers indicate the size of the standards (M) in nucleotides. (c) The DNA was isolated as indicated in Experimental procedures and 3 μg was electrophoresed. The numbers indicate the corresponding size of the standards (λDNA HindIII/EcoRI) in Kb.
Initially, the toxicity of RIPs to animal cells was attributed to their ability to inhibit protein synthesis leading to cell death, but there is an increasing body of evidence indicating that the main cause of RIP toxicity in these cells is their ability to induce apoptosis (Das et al., 2012). COLO 320 cells treated with BE27 exhibited the morphological features characteristic of apoptosis, strong dose‐dependent activation of the effector caspase 3/7 and the breakdown of the nuclear DNA into oligonucleosomal fragments (Iglesias et al., 2015). DNA from P. digitatum was isolated from the cultures shown in Fig. 4a and analysed to detect the presence of the oligonucleosomal fragments (Fig. 4c). Although a smear of degraded DNA was observed in BE27‐treated fungi compared with the untreated control, no internucleosomal cleavage was visible, suggesting that BE27 intoxication can occur by non‐apoptotic mechanisms.
In silico structural analysis of BE27
It has been suggested that a β‐hairpin present in domain 3 of the dicot type 1 RIPs may be necessary for the toxicity of such proteins (Di Maro et al., 2014). To ascertain the main structural characteristics that may be involved in the antifungal behaviour of BE27, a three‐dimensional structure was predicted by comparative modelling using several RIP crystal structures as templates. The selected best model was found to have a confidence score (C‐score) of 1.24, template modeling (Tm) score of 0.89 ± 0.07 and root‐mean‐square deviation (RMSD) of 3.3 ± 2.3 Å, which satisfied the range of parameters for molecular modelling. Figure 5a shows such a model that displays the characteristic fold of type 1 RIPs and the A chain of type 2 RIPs, in which the confluence of three domains creates the active site: N‐terminal domain 1, consisting of β‐strands and α‐helices; domain 2, characterized by α‐helices; and C‐terminal domain 3, formed by two α‐helices and two β‐strands (Di Maro et al., 2014). Although the models obtained by comparative modelling have a limited value and should be confirmed by X‐ray diffraction studies, we used such a model to compare the structure of BE27 with some of the well‐known antifungal peptide defensins (Fig. 5b–f). Plant defensins are small proteins of 45–54 amino acid residues which, despite their low level of amino acid sequence identity, present a conserved tertiary structure that consists of a triple‐stranded antiparallel β‐sheet and one α‐helix that are stabilized into a compact shape by three to five disulfide bridges (Lacerda et al., 1977). A β‐hairpin (the γ‐core motif) has been reported to be essential for membrane interactions allowing the entry of defensins into fungal cells (Lacerda et al., 1977; Sagaram et al., 2013). Such a β‐hairpin is preceded by the α‐helix. As shown in Fig. 5b–e, BE27 displayed the same motif as defensins next to its carboxy terminus, consisting of a helix of 10 amino acid residues linked to an opposing β‐hairpin by a loop of five residues. The β‐hairpin is composed of two antiparallel β‐strands of eight and six residues linked by a loop of five residues. This structure is almost identical to that of PAP‐S, a RIP isolated from Phytolacca americana L. (Fig. 5c). Figure 5b and 5c also show the helix and γ‐core motif from two representative antifungal defensins: SPE10 from Pschyrhizus erosus (L.) Urb. (Song et al., 2011) and MtDf4 from Medicago truncatula Gaertn. (Sagaram et al., 2013). Both defensins contain a helix of 11 residues and an opposing β‐hairpin containing two β‐strands (five and seven residues). These structures are characterized by the presence of several positive charges which, in the case of BE27, are supplied by the amino acids lysine (Lys)‐203, arginine (Arg)‐209, Lys‐210, Lys‐213, Lys‐214 and Lys‐229 (Fig. 5b). These amino acids confer to these structures a highly positively charged electrostatic surface (Fig. 5e), which is almost fully exposed in the BE27 structure (Fig. 5d). Such types of amino acids are required to interact, through electrostatic interactions, with the negatively charged sphingolipids of the fungal plasma membrane (Lacerda et al., 1977). BE27 also displays tryptophan (Trp)‐230, a hydrophobic residue conserved in antifungal defensins (Fig. 5b). This residue [phenylalanine (Phe) in other defensins] has been reported to be a key factor for antifungal action in SPE10 (Song et al., 2011). As shown in Fig. 5f, the α‐helix of BE27 assumes an amphipathic conformation with opposing nonpolar [Trp‐201, isoleucine (Ile)‐204, glycine (Gly)‐207, Ile‐208] and polar [serine (Ser)‐202, Lys‐203, Ser‐205, glutamic acid (Glu)‐206, Arg‐209, Lys‐210] faces oriented along the long axis of the helix. In addition, the polar face displays a high cationicity because of the presence of one Arg and two Lys. This type of conformation has been reported to permit an efficient interaction with biological membranes in antimicrobial peptides (Zelezetsky and Tossi, 2006). A similar conformation is present in the PAP α‐helix, whereas such a conformation is not evident in SPE10 or MtDef4 (Fig. 5f).
Figure 5.
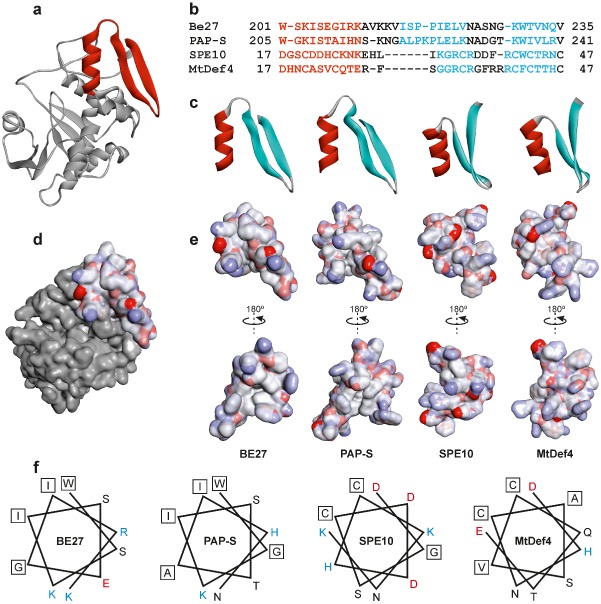
Structure of the α‐helix/γ‐core motif of BE27 compared with PAP‐S (pokeweed antiviral protein) and the defensins SPE10 and MtDef4. (a) Structure of BE27 indicating the position of the α‐helix/γ‐core motif from amino acid 201 to amino acid 235 (red ribbon). (b) Sequence alignments based on the three‐dimensional alignments using the program Expresso (Di Tommaso et al., 2011). Colouring of the alignments indicates the presence of α‐helix (red) or β‐sheet (cyan) secondary structures. (c) Ribbon representation of the α‐helix/γ‐core motif from BE27 expanding from amino acid 201 to amino acid 235 and their homologues from PAP‐S, SPE10 and MtDef4. The α‐helix and β‐strands are coloured in red and cyan, respectively. (d, e) Electrostatic surface of the α‐helix/γ‐core motif from BE27 localized in the whole protein (d) or compared with the same motif in PAP‐S, SPE10 and MtDef4 (e). Electrostatic potential is indicated in red (negative charge), white (neutral) and blue (positive charge). (f) Helical wheel drawing of the helices of BE27, PAP‐S, SPE10 and MtDef4. The amino acid charge is indicated in red (negative), black (neutral) and blue (positive). Hydrophobic amino acids are boxed.
Discussion
The type 1 RIP BE27 has been reported as a defence protein synthesized in response to virus infection whose mechanism of action lies in its RNA polynucleotide:adenosine glycosylase activity, thus conferring it with the ability to depurinate and destroy viral RNAs (Iglesias et al., 2005). However, other activities have been reported recently which might favour the resistance of beet against a broad variety of pathogens (Iglesias et al., 2015). The combined effect of these biological activities could result in a broad action against several types of pathogen, such as viruses, bacteria, fungi or insects. Because several studies have indicated that some RIPs protect plants from fungal infection (Park et al., 2002) and transgenic plants bearing RIP genes have been constructed that are resistant to fungi (Corrado et al., 2005; Huang et al., 2008; Qian et al., 2014), we have tested the ability of BE27 to arrest the growth of the fungus P. digitatum. The Penicillium genus includes necrotrophic fungi that colonize the wounds and grow in the inter‐ and intracellular spaces of plant tissues (de Capdeville et al., 2008). Crucial regulatory processes and protein–protein interactions take place in the apoplast during fungal infection (Doehlemann and Hemetsberger, 2013). Among the PR proteins which are secreted into the apoplast during basal resistance responses are chitinases, glucanases, thionins, osmotins, proteases and defensins (Doehlemann and Hemetsberger, 2013; van Loon et al., 2006). Here, we report that BE27 is another apoplastic protein able to enter and kill fungal cells, thus arresting the growth of the fungus. The mechanism of action seems to involve rRNA N‐glycosylase activity on the Penicillium major rRNA which inactivates irreversibly its ribosomes, thus inhibiting protein synthesis. By contrast, beet ribosomes are not sensitive to BE27 (Iglesias et al., 2008). This contrasts with data reported for PAP from Phytolacca americana L. leaves, which inactivates their ‘conspecific’ ribosomes (Prestle et al., 1992). PAP accumulates in the apoplastic space and has been postulated to be part of a general suicide strategy of some higher plants, rendering wounded tissue inefficient for the establishment of an infection by viruses or parasitic fungi (Park et al., 2004; Prestle et al., 1992). The different RIPs seem to display different antifungal capacities. It has been reported that ME, a type 1 RIP from Mirabilis expansa (Ruiz &Pav.) Standl., shows substantial inhibitory activity against Rhizoctonia solani growth, whereas the type 1 RIP saporin‐S6 (from Saponaria officinalis L.) does not display such activity, despite the fact that saporin‐S6 is more active against R. solani ribosomes (Park et al., 2002). ME was targeted to the surface of fungal cells and transferred into the cells; thus, ME caused ribosome depurination and subsequent fungal mortality. In contrast, saporin did not interact with fungal cells, correlating with its lack of antifungal activity (Park et al., 2002). It is worth mentioning that ME did not inhibit the growth of another fungus, Trichoderma reesei (Park et al., 2002), suggesting that RIP antifungal activity could be specific.
The DNA of P. digitatum cultures appeared to be degraded, but did not show internucleosomal cleavage on exposure to BE27, thus suggesting that the mechanism of fungal growth inhibition is ribosome inactivation, followed by necrotic cell death. This contrasts with the effect of some RIPs on insects, in which apoptosis in their gut cells seems to be the mechanism of toxicity (Shahidi‐Noghabi et al., 2011). However, we cannot exclude the possibility that the SOD activity reported for BE27 (Iglesias et al., 2015) may contribute to the toxicity towards fungal cells. SOD activity generates hydrogen peroxide, which can be toxic to fungi or may directly or indirectly stimulate plant defence responses (Doehlemann and Hemetsberger, 2013; van Loon et al., 2006). In this respect, it has been reported that SOD and other antioxidant enzymes from rice coordinately participate in the resistance to rice stripe virus (Hao et al., 2011), and that SOD transgenes confer resistance to the fungus Cercospora beticola in sugar beet (Tertivanidis et al., 2004).
Fluorescence microscopy of P. digitatum after incubation with BE27 conjugated with the fluorescent dye Cy3 indicates that BE27 is able to internalize and reach the cytoplasm of fungal cells. This is worth mentioning because type 1 RIPs internalize poorly and high concentrations are necessary to become toxic to intact cells. In fact, BE27 displays IC50 values of 2.2 × 10−6 and 4 × 10−7 m to COLO 320 cells (the most sensitive cellular culture) after 48 and 72 h of exposure, respectively (Iglesias et al., 2015). These concentrations are equivalent to 59 and 11 μg/mL, respectively, higher than that required to arrest P. digitatum growth. In order to study the structural peculiarities that may be involved in the antifungal behaviour of BE27, a three‐dimensional structure was predicted by comparative modelling and compared with PAP and the defensins SPE10 from Pschyrhizus erosus (Song et al., 2011) and MtDef4 from M. truncatula (Sagaram et al., 2013). Defensins are small proteins with a conserved signature of cysteines, which can form three to four disulfide bridges (Lacerda et al., 1977). An amino acid sequence alignment of antifungal defensins from plants shows that they do not present conservative amino acid sequences, except for the cysteine residues and a Gly residue (Lacerda et al., 1977). However, defensins present a well‐conserved three‐dimensional structure which forms one α‐helix and three antiparallel β‐sheets. These proteins can interact specifically with some negatively charged sphingolipids present at the cell membrane of pathogens, causing an increase in its permeabilization, leading to cell leakage and death by necrosis, or even to the transportation of these proteins to the intracellular environment, activating programmed cell death (Lacerda et al., 1977; Thevissen et al., 2003). The defensin specificity of binding to different sphingolipids or rafts containing sphingolipids and ergosterol is responsible for the disparity in toxicity of the different defensins against different fungi and the lack of toxicity towards plant cells (Thevissen et al., 2003). It has been reported that the carboxy‐terminal amino acid sequence is a major determinant of the antifungal activity of defensins (Sagaram et al., 2013). This sequence is characterized by the presence of two antiparallel β‐strands with an interposed loop and is preceded by an α‐helix. Such a structural motif is characterized by a highly positively charged electrostatic surface (Lacerda et al., 1977). In this study, we have identified a similar structural motif in the carboxy‐terminal amino acid sequence of BE27 and PAP. This motif is shared by all apoplastic type 1 RIPs from dicots, and absent in the cytoplasmic type 1 RIPs from monocots (Di Maro et al., 2014). In BE27, this motif also contains many positively charged amino acids required for the interaction with the negatively charged sphingolipid heads in the fungal membranes (Lacerda et al., 1977; Thevissen et al., 2003). However, the cysteines and disulfide bonds, a characteristic of all antifungal defensins, are absent in BE27. Because plant defensins lack a distinct hydrophobic core, the protein fold is stabilized primarily by disulfide bonds. This stabilization is achieved in type 1 RIPs by the presence of surrounding chains from the rest of the protein, particularly by the carboxy‐terminal extension that forms an interface with the rest of the protein. The amphipathic α‐helix of BE27 and PAP, characterized by a high cationicity of the polar face, might allow an efficient interaction with the fungal membrane, as has been reported for some antimicrobial peptides (Zelezetsky and Tossi, 2006). This might be necessary because PAP and, especially, BE27 present less cationic amino acids in their β‐hairpins than SPE10 or MtDef4. Such amino acids have been found to be relevant for interaction with the negatively charged sphingolipids of fungal plasma membranes (Lacerda et al., 1977; Sagaram et al., 2013; Thevissen et al., 2003). The insertion of BE27 into the lipid bilayer might be achieved by the presence of the β‐hairpin, because a well‐defined β‐hairpin conformation has been reported to be necessary for disruption of the lamellar structure of the lipid membrane and insertion into the bilayer of β‐hairpin antimicrobial peptides (Mani et al., 2005).
The involvement of the above‐mentioned structure in the entry and toxicity of BE27 and PAP in fungal cells is in accordance with studies reported on recombinant PAP mutants lacking several amino acids in their C‐terminus (Baykal and Tumer, 2007; Hudak et al., 2004; Hur et al., 1995; Tumer et al., 1997). Recombinant PAP is toxic to both transgenic tobacco (Tumer et al., 1997) and yeast (Baykal and Tumer, 2007; Hudak et al., 2004; Hur et al., 1995) bearing the PAP gene, because this protein inactivates the ribosomes of the host. By contrast, C‐terminal deletion mutants lacking a 25‐amino‐acid sequence (thus including part of the β‐hairpin) and displaying rRNA N‐glycosylase activity were not toxic to either host. This was a result of both a diminished stability of the mutant protein (Tumer et al., 1997) and a failure in their sorting from the endoplasmic reticulum to the cytosol through the reticulum membrane (Baykal and Tumer, 2007; Hudak et al., 2004; Hur et al., 1995; Tumer et al., 1997). However, this failure in the sorting to the cytosol was maintained by mutants having deletions just downstream of the β‐hairpin. Although the authors of these articles sympathized with the hypothesis that the sequence of the last 10 amino acids was responsible for targeting to the membranes, such a sequence is not present in BE27 because it possesses a six amino acid shorter C‐terminal sequence. A possible explanation is that the lack of these amino acids placed in the interface between the α‐helix/γ‐core motif and the rest of the protein might diminish the stability of the β‐hairpin structure in RIPs which, unlike defensins, lack the stabilizing disulfide bonds.
Further work will be conducted to study the in vitro and in vivo antifungal potential of BE27 and other RIPs against different fungi. The study of the toxicity of RIP mutants against fungal pathogens will clarify the role played by the C‐terminus in the interaction with the fungal plasma membrane.
Experimental Procedures
Materials
The sources of the chemicals used in this work have been indicated previously (Ferreras et al., 2011b) and most were obtained from Sigma‐Aldrich (St Louis, MO, USA). The strain of P. digitatum was isolated in our laboratory and typified by the Spanish Type Culture Collection (CECT), Valencia, Spain. Potato dextrose agar (PDA) medium (purchased from Sigma‐Aldrich) contains the following (per litre): 300 g potato infusion, 20 g dextrose, 15 g agar and 0.1 g chloramphenicol. Potato dextrose broth (PDB) medium (purchased from Sigma‐Aldrich) contains the following (per litre): 6 g potato extract and 20 g dextrose. BE27 was isolated from 300 g of sugar beet leaves collected in September from a crop field at Cobos de Cerrato (Palencia, Spain) following a procedure described previously (Iglesias et al., 2005).
Preparation of crude extracts from sugar beet leaves
All plants were grown in a growth chamber under conditions of 12 h light and 12 h dark at 20 °C. Treatment of laboratory‐grown sugar beets with either hydrogen peroxide or salicylic acid was carried out by spraying dilute solutions (5 mm) every 24 h over 3 days. The crude leaf extracts were obtained from leaves that were ground in a ceramic mortar with liquid nitrogen; 30–50 mg were transferred to an Eppendorf tube and extracted for 10 min with 10 volumes of 5 mm sodium phosphate (pH 7.5) buffer containing 140 mm NaCl. After centrifugation at 10 000 g at 5 °C for 10 min, the supernatant was removed and stored at −20 °C.
Production of conidia and mycelium
The fungus was cultured on PDA slants for 8–10 days in a 26 °C incubator. A conidial suspension of P. digitatum was prepared by gently scraping the culture surface using a sterile glass rod after the addition of 2–3 mL of 0.1% aqueous solution of Tween‐20. The spore suspension obtained was concentrated by centrifugation and dissolved in 0.1% aqueous solution of Tween‐20. The concentration of the conidial suspension was quantified with a haematocytometer and the suspension was stored at −20 °C. Mycelium was obtained by inoculation of 250‐mL portions of PDB in 1‐L sterile Erlenmeyer flasks with 0.5 mL of spore suspension. The flasks were incubated at 26 °C on a horizontal shaker (250 rpm). After growing P. digitatum for 4–5 days, the mycelium was harvested by filtration through filter paper under vacuum, washed with distilled water and stored at −20 °C.
Antifungal activity measurements
Growth inhibitory assays of BE27 against P. digitatum were performed in 96‐well microtitre plates. Conidia of P. digitatum (1000 spores/well) were incubated at 26 °C in 150 μL of PDB medium in the presence of different concentrations of BE27. Fungal growth was monitored spectrophotometrically using a microtitre plate Multiskan EX reader (Thermo Scientific, Waltham, MA, USA) and microscopically after 24, 28, 34, 45, 50, 71 and 78 h of incubation. Each experiment was performed in quadruplicate.
Mycelium for RNA and DNA extraction was prepared from cultures grown in 25‐mm plates containing 2 mL of PDB medium inoculated with 1333 spores in the absence or presence of 4 μg/mL BE27. The plates were incubated at 26 °C on a horizontal shaker (40 rpm). After the growth of P. digitatum for 5 days, the mycelium was harvested by filtration through filter paper under vacuum, extensively washed with sterile water, weighed and stored at −80 °C. The experiments were carried out with six plates and performed in duplicate.
Microscopy of P. digitatum mycelium
Conidia of P. digitatum were grown in vitro in PDB medium for 24 h before exposure to BE27 (5 μg/mL) for 24 h. After incubation, samples were stained with 0.1% (w/v) of the chitin dye CFW for 5 min in the dark. Finally, mycelium was washed, resuspended in 20% glycerol and visualized under a Zeiss Axiovert 200M fluorescence microscope (Zeiss, Jena, Germany).
Cy3‐B27 labelling and confocal laser scanning microscopy of P. digitatum samples
BE27 fluorescence labelling with Cy3 was performed using the Cy3 Mono‐Reactive Dye Pack from Amersham/GE‐Healthcare (Barcelona, Spain) following the manufacturer's instructions. Conidia of P. digitatum were grown in vitro in PDB medium for 24 h before exposure to Cy3‐BE27 (5 μg/mL) for 24 h. After washing, the samples were visualized with an LSM 510 META confocal microscope (Carl Zeiss, Jena, Germany).
rRNA N‐glycosylase activity of BE27 on yeast and P. digitatum ribosomes
Preparation of the 30 000 g (S30) supernatants from yeast and P. digitatum was performed as described elsewhere (Iglesias et al., 2015). Fresh baker's yeast (7.5 g) was ground in an unvitrified mortar precooled at −20 °C, with 15 g of alumina for 30 min at 4 °C, extracted with two volumes of buffer containing 10 mm Tris‐HCl pH 7.6, 10 mm magnesium acetate, 10 mm KCl and 6 mm 2‐mercaptoethanol, and centrifuged at 30 000 g for 20 min. This supernatant was designated S30 and was stored at −80 °C. The P. digitatum S30 supernatant was obtained from 15 g of mycelium as described for yeast. The rRNA N‐glycosylase activity of beetin was assayed in 100‐μL samples of S30 supernatants from yeast or P. digitatum, which were incubated with BE27 for 1 h at 30 °C. After treatment, the RNA was extracted with phenol and treated with aniline for 15 min at 23 °C. RNA samples were separated on a 5% urea–polyacrylamide gel, and stained with ethidium bromide (Iglesias et al., 2015).
rRNA N‐glycosylase activity in P. digitatum cultures
RNA from P. digitatum grown in the presence of BE27 was obtained from 15 mg of mycelium ground in a ceramic mortar with liquid nitrogen, using the RNA Plant Minikit (Qiagen, Madrid, Spain), according to the manufacturer's procedure. RNA samples were treated with aniline for 15 min at 23 °C, separated on a 5% urea–polyacrylamide gel and stained with ethidium bromide (Iglesias et al., 2015).
DNA fragmentation analysis in P. digitatum cultures
The DNA from P. digitatum grown in the presence of BE27 was obtained from 20 mg of mycelium ground in a ceramic mortar with liquid nitrogen; 15 mg were transferred to an Eppendorf tube and suspended in 100 μL of 1 m sorbitol containing 0.1 m ethylenediaminetetraacetic acid (EDTA), pH 7.4, 0.1% 2‐mercaptoethanol and 100 units of lyticase. After incubation at 30 °C for 1 h, the DNA was isolated following the instructions of the Genomic Prep Cells and Tissue DNA Isolation Kit (GE‐Healthcare, Barcelona, Spain). DNA (3 μg) electrophoresis was carried out in 1% agarose gels using TBE buffer (0.089 m Tris, 0.089 m boric acid, 2 mm EDTA, pH 8.0) at 50 V for 4 h. DNA was stained for 20 min with Gel Red (Biotium, Inc., Hayward, CA, USA) and visualized with an ultraviolet lamp.
Molecular modelling of BE27 and structural comparison
The complete amino acid sequence of BE27 (accession number AAS67266.1) (Iglesias et al., 2005) was downloaded from the National Center for Biotechnology Information (NCBI) sequence database (http://www.ncbi.nlm.nih.gov/protein/). Three‐dimensional structural modelling was carried out on the I‐TASSER server (Roy et al., 2010; http://zhanglab.ccmb.med.umich.edu/I‐TASSER). Protein structures 2q8w (PAP‐S1aci), 3ctk (bouganin), 1qci (PAP), 1wuc (bouganin), 2z4u (PD‐L4), 1ift (ricin A‐chain) and 1apa (abrin A‐chain) were chosen by I‐TASSER as templates in the modelling. The three‐dimensional structures of PAP‐S (accession code 1GIK), SPE10 (3PSM) and MtDef4 (2LR3) are available in the Protein Data Bank site (http://www.rcsb.org/pdb/home/home.do). Three‐dimensional alignments of BE27, Psd1 and MtDef4 with PAP‐S were performed using the program Expresso (Di Tommaso et al., 2011; http://tcoffee.crg.cat/apps/tcoffee/do:expresso). Study and graph representations of protein structures were performed with the aid of the Discovery Studio 3.5 suite (http://accelrys.com/).The helical wheels were generated using WHAT 2.0 (Saier et al., 2014; http://www.tcdb.org).
Other procedures
Chitinase activity was evaluated as described previously (Naher et al., 2012) using chitin as substrate and measuring the absorbance at 585 nm on addition of Ehrlich reagent (p‐dimethylaminobenzaldehyde dissolved in glacial acetic acid that contained HCl). Protein synthesis was performed with a coupled transcription–translation in vitro assay using a rabbit reticulocyte lysate system as described elsewhere (Ferreras et al., 2011b). Protein concentrations were determined using the spectrophotometric method of Kalb and Bernlohr (2014). The RIP content in the crude extracts was calculated as indicated by Barbieri et al. (2006).
Acknowledgements
This work was supported by grant BIO/VA39/14 (Consejería de Sanidad, Junta de Castilla y León) to L.C. C.G. was supported by an Eurotango II fellowship (Erasmus Mundus Program). We thank Judy Callaghan (Monash University, Melbourne, Australia) for correcting the manuscript.
References
- Barbieri, L. , Valbonesi, P. , Bonora, E. , Gorini, P. , Bolognesi, A. and Stirpe, F. (1997) Polynucleotide:adenosine glycosidase activity of ribosome‐inactivating proteins: effect on DNA, RNA and poly(A). Nucleic Acids Res. 25, 518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri, L. , Polito, L. , Bolognesi, A. , Ciani, M. , Pelosi, E. , Farini, V. , Jha, A.K. , Sharma, N. , Vivanco, J.M. , Chambery, A. , Parente, A. and Stirpe, F. (2006) Ribosome‐inactivating proteins in edible plants and purification and characterization of a new ribosome‐inactivating protein from Cucurbita moschata . Biochim. Biophys. Acta, 1760, 783–792. [DOI] [PubMed] [Google Scholar]
- Baykal, U. and Tumer, N.E. (2007) The C‐terminus of pokeweed antiviral protein has distinct roles in transport to the cytosol, ribosome depurination and cytotoxicity. Plant J. 49, 995–1007. [DOI] [PubMed] [Google Scholar]
- Becker, N. and Benhar, I. (2012) Antibody‐based immunotoxins for the treatment of cancer. Antibodies, 1, 39–69. [Google Scholar]
- Berglund, L. , Brunstedt, J. , Nielsen, K.K. , Chen, Z. , Mikkelsen, J.D. and Marcker, K.A. (1995) A proline‐rich chitinase from Beta vulgaris . Plant Mol. Biol. 27, 211–216. [DOI] [PubMed] [Google Scholar]
- de Capdeville, G. , Beer, S.V. , Wilson, C.L. and Aist, J.R. (2008) Some cellular correlates of hairpin induced resistence to blue mold of apples. Trop. Plant Pathol. 32, 103–113. [Google Scholar]
- Carzaniga, R. , Sinclair, L. , Fordham‐Skelton, A.P. , Harris, N. and Croy, R.R.D. (1994) Cellular and subcellular distribution of saporins, type‐1 ribosome‐inactivating proteins, in soapwort (Saponaria officinalis L.). Planta, 194, 461–470. [Google Scholar]
- Chaudhry, B. , Muller‐Uri, F. , Cameron‐Mills, V. , Gough, S. , Simpson, D. , Skriver, K. and Mundy, J. (1994) The barley 60 kDa jasmonate‐induced protein (JIP60) is a novel ribosome‐inactivating protein. Plant J. 6, 815–824. [DOI] [PubMed] [Google Scholar]
- Citores, L. , Iglesias, R. and Ferreras, J.M. (2013) Ribosome inactivating proteins from plants: biological properties and their use in experimental therapy In: Antitumor Potential and Other Emerging Medicinal Properties of Natural Compounds (Fang E.F. and Ng T.B., eds), pp. 127–143. Dordrecht: Springer. [Google Scholar]
- Corrado, G. , Bovi, P.D. , Ciliento, R. , Gaudio, L. , Di Maro, A. , Aceto, S. , Lorito, M. and Rao, R. (2005) Inducible expression of a Phytolacca heterotepala ribosome‐inactivating protein leads to enhanced resistance against major fungal pathogens in tobacco. Phytopathology, 95, 206–215. [DOI] [PubMed] [Google Scholar]
- Das, M.K. , Sharma, R.S. and Mishra, V. (2012) Induction of apoptosis by ribosome inactivating proteins: importance of N‐glycosidase activity. Appl. Biochem. Biotechnol. 166, 1552–1561. [DOI] [PubMed] [Google Scholar]
- Di Cola, A. , Marcozzi, G. , Balestrini, R. and Spano, R. (1999) Localization of the type I ribosome inactivating protein, luffin, in adult and embryonic tissues of Luffa cylindrica L. Roem. J. Exp. Bot. 50, 573–579. [Google Scholar]
- Di Maro, A. , Citores, L. , Russo, R. , Iglesias, R. and Ferreras, J.M. (2014) Sequence comparison and phylogenetic analysis by the Maximum Likelihood method of ribosome‐inactivating proteins from angiosperms. Plant Mol. Biol. 85, 575–588. [DOI] [PubMed] [Google Scholar]
- Di Tommaso, P. , Moretti, S. , Xenarios, I. , Orobitg, M. , Montanyola, A. , Chang, J.M. , Taly, J.F. and Notredame, C. (2011) T‐Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39, W13–W17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlemann, G. and Hemetsberger, C. (2013) Apoplastic immunity and its suppression by filamentous plant pathogens. New Phytol. 198, 1001–1016. [DOI] [PubMed] [Google Scholar]
- Dowd, P.F. , Johnson, E.T. and Price, N.P. (2012) Enhanced pest resistance of maize leaves expressing monocot crop plant‐derived ribosome‐inactivating protein and agglutinin. J. Agric. Food Chem. 60, 10 768–10 775. [DOI] [PubMed] [Google Scholar]
- Engelberth, J. , Contreras, C.F. and Viswanathan, S. (2012) Transcriptional analysis of distant signaling induced by insect elicitors and mechanical wounding in Zea mays . PLoS ONE, 7, e34855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, E.F. , Ng, T.B. , Shaw, P.C. and Wong, R.N. (2011) Recent progress in medicinal investigations on trichosanthin and other ribosome inactivating proteins from the plant genus Trichosanthes . Curr. Med. Chem. 18, 4410–4417. [DOI] [PubMed] [Google Scholar]
- Ferreras, J.M. , Citores, L. , Iglesias, R. , Jimenez, P. and Girbes, T. (2011a) Use of ribosome‐inactivating proteins from Sambucus for the construction of immunotoxins and conjugates for cancer therapy. Toxins, 3, 420–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreras, J.M. , Citores, L. , Iglesias, R. , Souza, A.M. , Jimenez, P. , Gayoso, M.J. and Girbes, T. (2011b) Occurrence of the type two ribosome‐inactivating protein nigrin b in elderberry (Sambucus nigra L.) bark. Food Res. Int. 44, 2798–2805. [Google Scholar]
- Frisvad, J.C. and Samson, R.A. (2004) Polyphasic taxonomy of Penicillium subgenus Penicillium. A guide to identification of the food and air‐borne terverticillate Penicillia and their mycotoxins. Stud. Mycol. 49, 1–174. [Google Scholar]
- Girbes, T. and Ferreras, J.M. (1998) Ribosome‐inactivating proteins from plants. Recent Res. Devel. Agric. Biol. Chem. 2, 1–16. [Google Scholar]
- Girbes, T. , de Torre, C. , Iglesias, R. , Ferreras, J.M. and Mendez, E. (1996) RIP for viruses. Nature, 379, 777–778.8587600 [Google Scholar]
- Girbes, T. , Ferreras, J.M. , Arias, F.J. and Stirpe, F. (2004) Description, distribution, activity and phylogenetic relationship of ribosome‐inactivating proteins in plants, fungi and bacteria. Mini Rev. Med. Chem. 4, 461–476. [DOI] [PubMed] [Google Scholar]
- Hao, Z. , Wang, L. , He, Y. , Liang, J. and Tao, R. (2011) Expression of defense genes and activities of antioxidant enzymes in rice resistance to rice stripe virus and small brown planthopper. Plant Physiol. Biochem. 49, 744–751. [DOI] [PubMed] [Google Scholar]
- Huang, M. , Hou, P. , Wei, Q. , Xu, Y. and Chen, A. (2008) A ribosome‐inactivating protein (curcin 2) induced from Jatropha curcas can reduce viral and fungal infection in transgenic tobacco. Plant Growth Regul. 54, 115–123. [Google Scholar]
- Hudak, K.A. , Parikh, B.A. , Di, R. , Baricevic, M. , Santana, M. , Seskar, M. and Tumer, N.E. (2004) Generation of pokeweed antiviral protein mutations in Saccharomyces cerevisiae: evidence that ribosome depurination is not sufficient for cytotoxicity. Nucleic Acids Res. 32, 4244–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur, Y. , Hwang, D.J. , Zoubenko, O. , Coetzer, C. , Uckun, F.M. and Tumer, N.E. (1995) Isolation and characterization of pokeweed antiviral protein mutations in Saccharomyces cerevisiae: identification of residues important for toxicity. Proc. Natl. Acad. Sci. USA, 92, 8448–8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias, R. , Perez, Y. , de Torre, C. , Ferreras, J.M. , Antolin, P. , Jimenez, P. , Rojo, M.A. , Mendez, E. and Girbes, T. (2005) Molecular characterization and systemic induction of single‐chain ribosome‐inactivating proteins (RIPs) in sugar beet (Beta vulgaris) leaves. J. Exp. Bot. 56, 1675–1684. [DOI] [PubMed] [Google Scholar]
- Iglesias, R. , Perez, Y. , Citores, L. , Ferreras, J.M. , Mendez, E. and Girbes, T. (2008) Elicitor‐dependent expression of the ribosome‐inactivating protein beetin is developmentally regulated. J. Exp. Bot. 59, 1215–1223. [DOI] [PubMed] [Google Scholar]
- Iglesias, R. , Citores, L. , Di Maro, A. and Ferreras, J.M. (2015) Biological activities of the antiviral protein BE27 from sugar beet (Beta vulgaris L.). Planta, 241, 421–433. [DOI] [PubMed] [Google Scholar]
- Kalb, V.F. and Bernlohr, R.W. (1977) A new spectrophotometric assay for protein in cell extracts. Anal. Biochem. 82, 362–371. [DOI] [PubMed] [Google Scholar]
- Lacerda, A.F. , Vasconcelos, E.A. , Pelegrini, P.B. and de Sa, M.F. (2014) defensins and their role in plant defense. Front. Microbiol. 5, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzanova, C. , Torri, A. , Motto, M. and Balconi, C. (2011) Characterization of the maize b‐32 ribosome inactivating protein and its interaction with fungal pathogen development. Maydica, 56, 83–93. [Google Scholar]
- van Loon, L.C. , Rep, M. and Pieterse, C.M. (2006) Significance of inducible defense‐related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. [DOI] [PubMed] [Google Scholar]
- Lopez‐Millan, A.F. , Morales, F. , Abadia, A. and Abadia, J. (2000) Effects of iron deficiency on the composition of the leaf apoplastic fluid and xylem sap in sugar beet. Implications for iron and carbon transport. Plant Physiol. 124, 873–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord, J.M. and Spooner, R.A. (2011) Ricin trafficking in plant and mammalian cells. Toxins, 3, 787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani, R. , Waring, A.J. , Lehrer, R.I. and Hong, M. (2005) Membrane‐disruptive abilities of beta‐hairpin antimicrobial peptides correlate with conformation and activity: a 31P and 1H NMR study. Biochim. Biophys. Acta, 1716, 11–18. [DOI] [PubMed] [Google Scholar]
- Naher, L. , Tan, S.G. , Yusuf, U.K. , Ho, C.L. and Siddiquee, S. (2012) Activities of chitinase enzymes in the oil palm (Elaeis guineensis Jacq.) in interactions with pathogenic and non‐pathogenic fungi. Plant Omics J. 5, 333–336. [Google Scholar]
- Pan, W.L. , Wong, J.H. , Fang, E.F. , Chan, Y.S. , Ng, T.B. and Cheung, R.C. (2014) Preferential cytotoxicity of the type I ribosome inactivating protein alpha‐momorcharin on human nasopharyngeal carcinoma cells under normoxia and hypoxia. Biochem. Pharmacol. 89, 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parente, A. , Conforto, B. , Di Maro, A. , Chambery, A. , De Luca, P. , Bolognesi, A. , Iriti, M. and Faoro, F. (2008) Type 1 ribosome‐inactivating proteins from Phytolacca dioica L. leaves: differential seasonal and age expression, and cellular localization. Planta, 228, 963–975. [DOI] [PubMed] [Google Scholar]
- Park, S.W. , Stevens, N.M. and Vivanco, J.M. (2002) Enzymatic specificity of three ribosome‐inactivating proteins against fungal ribosomes, and correlation with antifungal activity. Planta, 216, 227–234. [DOI] [PubMed] [Google Scholar]
- Park, S.W. , Vepachedu, R. , Sharma, N. and Vivanco, J.M. (2004) Ribosome‐inactivating proteins in plant biology. Planta, 219, 1093–1096. [DOI] [PubMed] [Google Scholar]
- Polito, L. , Bortolotti, M. , Pedrazzi, M. and Bolognesi, A. (2011) Immunotoxins and other conjugates containing saporin‐s6 for cancer therapy. Toxins, 3, 697–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestle, J. , Schonfelder, M. , Adam, G. and Mundry, K.W. (1992) Type 1 ribosome‐inactivating proteins depurinate plant 25S rRNA without species specificity. Nucleic Acids Res. 20, 3179–3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri, M. , Kaur, I. , Perugini, M.A. and Gupta, R.C. (2012) Ribosome‐inactivating proteins: current status and biomedical applications. Drug Discov. Today, 17, 774–783. [DOI] [PubMed] [Google Scholar]
- Qian, Q. , Huang, L. , Yi, R. , Wang, S. and Ding, Y. (2014) Enhanced resistance to blast fungus in rice (Oryza sativa L.) by expressing the ribosome‐inactivating protein alpha‐momorcharin. Plant Sci. 217–218, 1–7. [DOI] [PubMed] [Google Scholar]
- Qin, X. , Zheng, X. , Shao, C. , Gao, J. , Jiang, L. , Zhu, X. , Yan, F. , Tang, L. , Xu, Y. and Chen, F. (2009) Stress‐induced curcin‐L promoter in leaves of Jatropha curcas L. and characterization in transgenic tobacco. Planta, 230, 387–395. [DOI] [PubMed] [Google Scholar]
- Rippmann, J.F. , Michalowski, C.B. , Nelson, D.E. and Bohnert, H.J. (1997) Induction of a ribosome‐inactivating protein upon environmental stress. Plant Mol. Biol. 35, 701–709. [DOI] [PubMed] [Google Scholar]
- Roy, A. , Kucukural, A. and Zhang, Y. (2010) I‐TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagaram, U.S. , El‐Mounadi, K. , Buchko, G.W. , Berg, H.R. , Kaur, J. , Pandurangi, R.S. , Smith, T.J. and Shah, D.M. (2013) Structural and functional studies of a phosphatidic acid‐binding antifungal plant defensin MtDef4: identification of an RGFRRR motif governing fungal cell entry. PLoS ONE, 8, e82485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier, M.H. , Reddy, V.S. , Tamang, D.G. and Vastermark, A. (2014) The transporter classification database. Nucleic Acids Res. 42, D251–D258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahidi‐Noghabi, S. , Van Damme, E.J. , De Vos, W.H. and Smagghe, G. (2011) Internalization of Sambucus nigra agglutinins I and II in insect midgut CF‐203 cells. Arch. Insect Biochem. Physiol. 76, 211–222. [DOI] [PubMed] [Google Scholar]
- Shih, N.R. , McDonald, K.A. , Jackman, A.P. , Girbes, T. and Iglesias, R. (1997) Bifunctional plant defence enzymes with chitinase and ribosome inactivating activities from Trichosanthes kirilowii cell cultures. Plant Sci. 130, 145–150. [Google Scholar]
- Song, S.K. , Choi, Y. , Moon, Y.H. , Kim, S.G. , Choi, Y.D. and Lee, J.S. (2000) Systemic induction of a Phytolacca insularis antiviral protein gene by mechanical wounding, jasmonic acid, and abscisic acid. Plant Mol. Biol. 43, 439–450. [DOI] [PubMed] [Google Scholar]
- Song, X. , Zhang, M. , Zhou, Z. and Gong, W. (2011) Ultra‐high resolution crystal structure of a dimeric defensin SPE10. FEBS Lett. 585, 300–306. [DOI] [PubMed] [Google Scholar]
- Tan, Y.C. , Yeoh, K.A. , Wong, M.Y. and Ho, C.L. (2013) Expression profiles of putative defence‐related proteins in oil palm (Elaeis guineensis) colonized by Ganoderma boninense . J. Plant Physiol. 170, 1455–1460. [DOI] [PubMed] [Google Scholar]
- Tertivanidis, K. , Goudoula, C. , Vasilikiotis, C. , Hassiotou, E. , Perl‐Treves, R. and Tsaftaris, A. (2004) Superoxide dismutase transgenes in sugarbeets confer resistance to oxidative agents and the fungus C. beticola . Transgenic Res. 13, 225–233. [DOI] [PubMed] [Google Scholar]
- Thevissen, K. , Ferket, K.K. , Francois, I.E. and Cammue, B.P. (2003) Interactions of antifungal plant defensins with fungal membrane components. Peptides, 24, 1705–1712. [DOI] [PubMed] [Google Scholar]
- Tumer, N.E. , Hwang, D.J. and Bonness, M. (1997) C‐terminal deletion mutant of pokeweed antiviral protein inhibits viral infection but does not depurinate host ribosomes. Proc. Natl. Acad. Sci. USA, 94, 3866–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter, H. , Robinson, D.G. and Heldt, H.W. (1994) Subcellular volumes and metabolite concentrations in spinach leaves. Planta, 193, 530–535. [Google Scholar]
- Yoshinari, S. , Yokota, S. , Sawamoto, H. , Koresawa, S. , Tamura, M. and Endo, Y. (1996) Purification, characterization and subcellular localization of a type‐1 ribosome‐inactivating protein from the sarcocarp of Cucurbita pepo . Eur. J. Biochem. 242, 585–591. [DOI] [PubMed] [Google Scholar]
- Yoshinari, S. , Koresawa, S. , Yokota, S. , Sawamoto, H. , Tamura, M. and Endo, Y. (1997) Gypsophilin, a new type 1 ribosome‐inactivating protein from Gypsophila elegans: purification, enzymatic characterization, and subcellular localization. Biosci. Biotechnol. Biochem. 61, 324–331. [DOI] [PubMed] [Google Scholar]
- Zelezetsky, I. and Tossi, A. (2006) Alpha‐helical antimicrobial peptides—using a sequence template to guide structure–activity relationship studies. Biochim. Biophys. Acta, 1758, 1436–1449. [DOI] [PubMed] [Google Scholar]


