Figure 3.
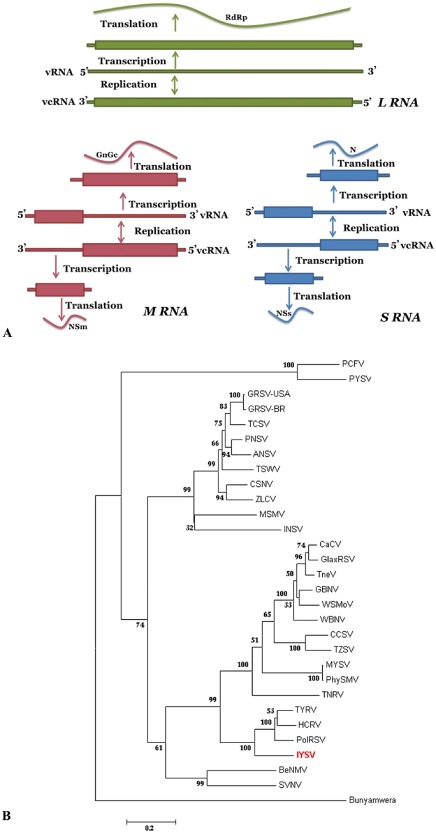
(A) Schematic representation of the genome organization and replication strategy of tospoviruses, showing the three RNAs: large (L), medium (M) and small (S). The rectangular boxes indicate the proteins coded. (B) Phylogeny based on amino acid sequences of nucleocapsid protein of known tospoviruses. The tospoviruses fall into two major groups based on their prevalence and distribution: Eurasia consisting of Europe, Asia and Oceania; the Americas consisting of North and South America. The representative gene sequence was obtained from the National Center for Biotechnology Information (NCBI) GenBank. Virus acronyms used in TREE are: Groundnut ringspot virus (GRSV); Groundnut bud necrosis virus (GBNV); Impatiens necrotic spot virus (INSV); Iris yellow spot virus (IYSV); Peanut yellow spot virus (PYSV); Polygonum ringspot virus (PolRSV); Tomato chlorotic spot virus (TCSV); Tomato spotted wilt virus (TSWV); Watermelon bud necrosis virus (WBNV); Zucchini lethal chlorosis virus (ZLCV); Alstroemeria necrotic streak virus (ANSV); Bean necrotic mosaic virus (BeNMV); Calla lily chlorotic spot virus (CCSV); Capsicum chlorosis virus (CaCV); Chrysanthemum stem necrosis virus (CSNV); Groundnut ringspot virus‐USA (GRSV‐USA); Gloxinia tospovirus (GlaxRSV); Hippeastrum chlorotic ringspot virus (HCRV); Melon severe mosaic virus (MSMV); Melon yellowspot virus (MYSV); Pepper necrotic spot virus (PNSV); Peanut chlorotic fanspot virus (PCFV); Physalis silver mottle virus (PhySMV); Soybean vein necrosis virus (SVNV); Tomato necrosis virus (TNeV); Tomato necrotic ringspot virus (TNRV); Tomato yellow fruit ring virus (TYFRV); Tomato zonate spot virus (TZSV); Watermelon silver mottle virus (WSMoV).
