Figure 4.
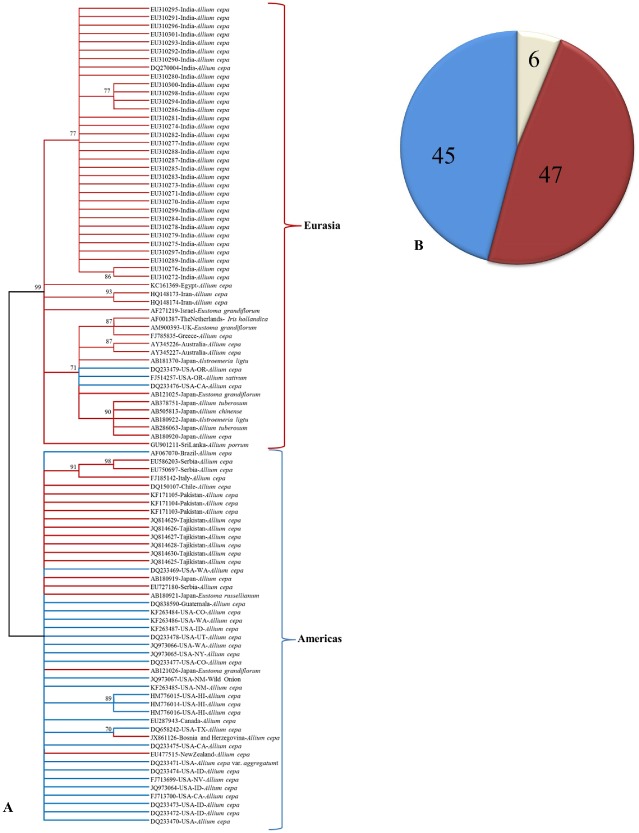
(A) Phylogeny based on amino acid sequences of the nucleocapsid protein of Iris yellow spot virus (IYSV) available in GenBank. The evolutionary history was inferred using the unweighted pair group method with arithmetic average (UPGMA). The optimal tree with the sum of branch length = 0.76736606 is shown. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. The analysis involved 99 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 270 positions in the final dataset. Evolutionary analyses were conducted in MEGA6. (B) Genotyping of IYSV accessions based on in silico restriction fragment length polymorphism (RFLP) simulation of nucleocapsid (N) gene (percentage of accessions under various genotypes).
