Summary
The resistance to a set of strains of Cucumber mosaic virus (CMV) in the melon accession PI 161375, cultivar ‘Songwhan Charmi’, is dependent on one recessive gene, cmv1, which confers total resistance, whereas a second set of strains is able to overcome it. We tested 11 strains of CMV subgroups I and II in the melon line SC12‐1‐99, which carries the gene cmv1, and showed that this gene confers resistance to strains of subgroup II only and that restriction is not related to either viral replication or cell‐to‐cell movement. This is the first time that a resistant trait has been correlated with CMV subgroups. Using infectious clones of the CMV strains LS (subgroup II) and FNY (subgroup I), we generated rearrangements and viral chimaeras between both strains and established that the determinant of virulence against the gene cmv1 resides in the first 209 amino acids of the movement protein, as this region from FNY is sufficient to confer virulence to the LS clone in the line SC12‐1‐99. A comparison of the sequences of the strains of both subgroups in this region shows that there are five main positions shared by all strains of subgroup II, which are different from those of subgroup I. Site‐directed mutagenesis of the CMV‐LS clone to substitute these residues for those of CMV‐FNY revealed that a combination of four of these changes [the group 64–68 (SNNLL to HGRIA), and the point mutations R81C, G171T and A195I] was required for a complete gain of function of the LS MP in the resistant melon plant.
Keywords: cmv1, Cucumber mosaic virus, melon, movement protein, resistance, virulence determinant
Introduction
To be able to infect a plant, viruses must interact with the host products by means of their own virulence factors. This interaction allows the virus to start or continue its infectious cycle and to develop disease. For genomes as tightly packed as those of viruses, it is important and convenient to be able to use all of their gene products, even in multiple functions, to facilitate the invasion of the host. Consequently, virulence factors can be either parts of the viral RNA itself (Albiach‐Marti et al., 2010; Díaz et al., 2004) or gene products of the virus (Choi et al., 2013; Decroocq et al., 2009; Mansilla et al., 2009; Mochizuki and Ohki, 2011). Cucumber mosaic virus (CMV) is able to infect more than 1200 species worldwide from over 100 families (Edwardson and Christie, 1991). To infect such a number of species, CMV has evolved to develop a vast genetic diversity (Palukaitis and Garcia‐Arenal, 2003). CMV isolates belong to two subgroups, I and II, defined by their serological and biological properties, and showing 73%–78% sequence similarity among them. Subgroup I is also divided into groups IA and IB, with 92%–94% sequence similarity between them (Roossinck, 2001). CMV has three genomic and two subgenomic RNAs. RNA1 encodes the 1a protein with two domains: a methyltransferase and a helicase. RNA2 encodes the 2a protein, an RNA‐dependent RNA polymerase, which, together with the 1a protein, is involved in the replication of the viral RNA. RNA2 also encodes the 2b protein, a suppressor of RNA silencing, which is also involved in long‐distance movement. RNA3 is bi‐cistronic and encodes the 3a protein, which is the movement protein (MP), and the 3b protein, the coat protein (CP). The 2b and 3b proteins are translated from two subgenomic RNAs, RNA4A and RNA4, respectively (Palukaitis and Garcia‐Arenal, 2003). CMV has also evolved the capacity to use as determinants of virulence all protein products encoded by its genome, depending on the host. All CMV proteins have been found to act as determinants of virulence in different systems (for a review, see Mochizuki and Ohki, 2011). The 1a protein is involved in the production of necrosis in some species of Nicotiana (Divéki et al., 2004) and can be methylated by a host protein, increasing the spread of CMV (Kim et al., 2008). In pepper, the helicase domain determines systemic infection in Cmr1‐mediated resistant plants (Kang et al., 2012). The 2a protein is related to the production of necrotic lesions in cowpea (Kim and Palukaitis, 1997) and also determines the systemic symptoms in squash by the facilitation of host‐specific viral movement (Choi et al., 2005). The 2b protein determines symptoms by interfering with the microRNA pathways (Diaz‐Pendon et al., 2007; Lewsey et al., 2007). The 3b protein (CP) is the most frequent virulence determinant reported in CMV in different host systems, in both monocots (Ryu et al., 1998) and dicots (Ryabov et al., 1999; Salánki et al., 2011; Takahashi et al., 2001; Taliansky and García‐Arenal, 1995; Thompson et al., 2006; Wong et al., 1999), and often by affecting cell‐to‐cell movement and/or systemic spread. In cucumber, a role for the CP as the determinant for long‐distance movement was reported using exchanges between two cucumoviruses. Tomato aspermy virus (TAV) is not able to infect cucumber systemically, whereas CMV can. Exchanges between TAV and CMV showed that only the CP of CMV could complement the lack of movement of TAV in cucumber (Taliansky and García‐Arenal, 1995). The 3a protein is the CMV product less frequently reported as the determinant of virulence in different systems. It has been related to the appearance of cyclic symptoms in tobacco (Gal‐On et al., 1996), but also to systemic infection in squash (Choi et al., 2005) and other cucurbits (Kaplan et al., 1997), and determines infection in soybean (Hong et al., 2007). Mutants generated in several domains of the MP affect the pathogenicity in different hosts, enabling the virus to invade systemically species or cultivars previously not allowed, or presenting altered symptoms in normal hosts (Kaplan et al., 1997; Li et al., 2001), indicating that different domains of the protein are involved in the determination of the infection in different hosts.
In the melon accession ‘Songwhan Charmi’ PI 161375 (SC), the resistance to CMV is oligogenic and recessive (Karchi et al., 1975). One of the genes involved, cmv1, confers resistance against CMV strains P9 and P104.82 (Essafi et al., 2009), but not to strains TL and M6. For these strains, the contribution of two other quantitative trait loci (QTLs), cmqw3.1 and cmqw10.1, which must act together with cmv1, is necessary to abort the infection (Guiu‐Aragonés et al., 2014). In this report, we show that strains of subgroup I are able to overcome cmv1‐mediated resistance, which is only effective against strains of subgroup II. We also show that this resistance does not involve impairment of virus replication and cell‐to‐cell movement. As a first step to understand the interactions taking place between CMV and the resistant melon line, we analysed the differences between the two subgroups of strains and identified the MP as the determinant of virulence. Moreover, we also demonstrated that a combination of three amino acids and a cluster of five amino acids of the MP, common to all tested strains of subgroup I, was sufficient to overcome cmv1‐mediated resistance.
Results
cmv1 confers resistance to strains of subgroup II only
As cmv1 confers total resistance to some strains, but not to others, we tested 11 CMV strains of both subgroups I and II for their ability to overcome cmv1‐mediated resistance to determine whether there was a correlation of the virulent phenotype with any of the subgroups. Plants of the resistant line SC12‐1‐99, carrying cmv1, were inoculated with sap from strains FNY, I17F, Ri‐8, M6 and Y (belonging to subgroup IA), Pl2 and Co1 (belonging to subgroup IB) and p104.82, WL, LS and NG (belonging to subgroup II). All subgroup I (IA and IB) strains were infectious in SC12‐1‐99, the line carrying cmv1, starting to produce symptoms at 7 days post‐inoculation (dpi) and developing a full systemic infection, whereas type II strains were unable to infect systemically this resistant line. The results are shown only for CMV‐LS and CMV‐FNY, the two most commonly experimentally used CMV strains (Fig. 1). The plants inoculated with CMV‐FNY showed the systemic infection typical of CMV, whereas the resistant line inoculated with CMV‐LS was completely asymptomatic. Accordingly, reverse transcriptase‐polymerase chain reaction (RT‐PCR) analysis was able to detect CMV‐FNY in SC12‐1‐99 plants, but failed to detect CMV‐LS in non‐inoculated leaves of this resistant line (Fig. 1b). To test whether this resistance to strains of subgroup II restricts viral replication or movement from the sites of inoculation, we analysed viral accumulation in cotyledons of PS and SC12‐1‐99 melon plants inoculated with either CMV‐FNY, as a member of subgroup I, or CMV‐LS, as a member of subgroup II. Northern blot analysis at 2 and 5 dpi revealed that CMV‐LS RNA accumulation did not differ significantly between the two melon lines and that, at 5 dpi, the virus accumulated at higher rates (Fig. 1c). As expected, CMV‐FNY RNA accumulated in inoculated leaves of resistant and susceptible melon lines. This indicated that replication and cell‐to‐cell movement of CMV‐LS were not restricted in the resistant SC12‐1‐99 line, despite the fact that systemic spread did not occur in these plants.
Figure 1.
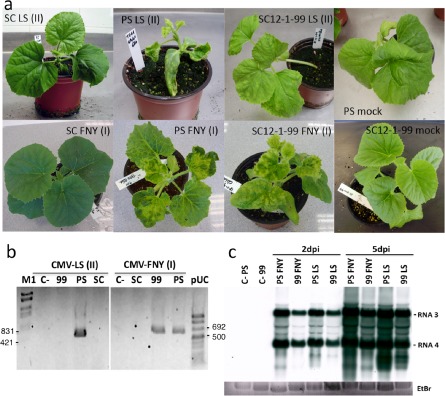
Symptoms and virus detection in melon genotypes inoculated with Cucumber mosaic virus (CMV)‐LS (subgroup II) and CMV‐FNY (subgroup I). (a) Symptoms in melon SC (Songwhan Charmi) harbouring the cmv1 allele, PS (Piel de Sapo) harbouring the CMV1 allele and the SC12‐1‐99 line harbouring the cmv1 allele, inoculated with CMV‐LS and CMV‐FNY. Photographs were taken at 20 days post‐inoculation (dpi). (b) Reverse transcriptase‐polymerase chain reaction (RT‐PCR) at 20 dpi for virus detection in the upper leaves of melon genotypes Songwhan Charmi (SC), Piel de Sapo (PS) and SC12‐1‐99 (99); C‐, mock‐inoculated plant; M1, Lambda Marker (EcoRI, HindIII); pUC, pUC Mix Marker 8 (Fermentas, Waltham, MA, USA). (c) Northern blot showing virus accumulation in inoculated cotyledons of Piel de Sapo (PS) and SC12‐1‐99 (99) at 2 and 5 dpi.
Therefore, there was a correlation between virulence in the cmv1 line and subgroup I, indicating that the gene cmv1 confers resistance to strains of subgroup II only, and that this resistance is not related to the impairment of viral accumulation in the inoculated cotyledon.
The MP is the determinant of virulence in cmv1‐mediated resistance
To account for this correlation between subgroups of CMV and cmv1‐mediated resistance, the two subgroups must differ with regard to the viral factor that determines the virulence in the line carrying cmv1. The molecular clones of the strains LS (subgroup II) and FNY (subgroup IA) are available for inoculation and manipulation (Rizzo and Palukaitis, 1990; Zhang et al., 1994) and were used to produce combinations between the three independent RNAs of each strain. ‘In vitro’‐transcribed RNAs were combined to produce all pseudo‐recombinants (Fig. 2a) and inoculated onto Nicotiana benthamiana plants to produce a high viral yield for subsequent inoculation into melon plants. As shown in Fig. 2, the only combination that caused virulence in the line SC12‐1‐99 (cmv1) carried RNA3 from FNY, and RNA1 and RNA2 from LS. The corresponding complementary combination (RNA3 from LS, and RNA1 and RNA2 from FNY) was unable to infect the resistant line SC12‐1‐99. All combinations shown in Fig. 2 were infectious in PS and none in SC; therefore, the fitness of the resulting combinations was not compromised. RT‐PCR analysis using primers specific to the unique RNA confirmed the presence of the correct combination in the upper leaves (Fig. 2b). Therefore, the determinant that confers virulence to CMV‐FNY (subgroup I) in the presence of the gene cmv1 was encoded in RNA3.
Figure 2.
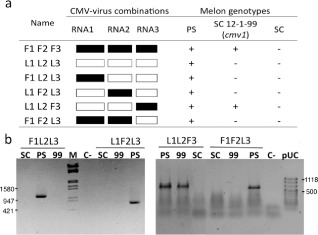
The determinant of virulence is in RNA3. (a) Schematic representation of the genome structure of pseudo‐recombinants between Cucumber mosaic virus (CMV)‐FNY (subgroup I) and CMV‐LS (subgroup II). CMV‐FNY (F1 F2 F3) is represented in black and CMV‐LS (L1 L2 L3) is represented in white. Results from both reverse transcriptase‐polymerase chain reaction (RT‐PCR) and visual symptoms of virus are indicated as infected (+) or non‐infected (–). (b) Infectivity of pseudo‐recombinants tested by RT‐PCR of the upper leaves at 20 days post‐inoculation (dpi) in melon genotypes Songwhan Charmi (SC) harbouring the cmv1 allele, Piel de Sapo (PS) harbouring the CMV1 allele and SC12‐1‐99 (99) harbouring the cmv1 allele. M, Lambda marker (EcoRI, HindIII); pUC, pUC Mix Marker 8 (Fermentas, Waltham, MA, USA); C‐, mock‐inoculated plant.
To further determine which element of RNA3 was responsible for the phenotype, we generated chimaeras exchanging either the three untranslated regions [5′UTR, intergenic region (IGR) and 3′UTR] independently, or the two open reading frames (ORFs), MP or CP genes. To assign a clear role to each region, the chimaeras were designed to exchange only the exact nucleotides corresponding to each region. As shown in Fig. 3a, a virulent phenotype was only produced when the MP of FNY was present. Even when only the MP of FNY (subgroup I) was present in the background of LS (subgroup II) [clone L3(MPFNY)], the virus was able to overcome the resistance. It produced the typical mosaic symptoms of CMV and was detected by RT‐PCR in systemically infected leaves using primers specific for the recombinant chimaera.
Figure 3.
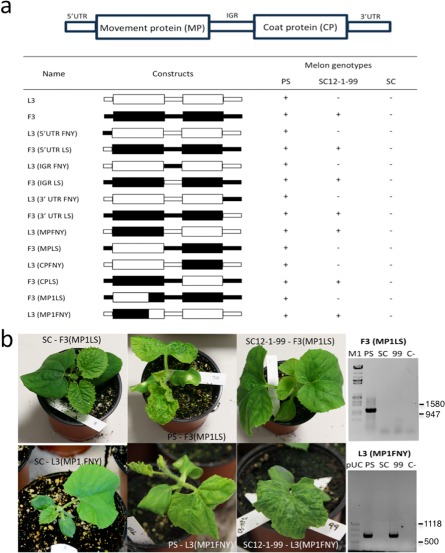
The determinant of virulence is in the movement protein (MP). (a) Chimaeric virus between Cucumber mosaic virus (CMV)‐FNY (subgroup I, represented in black) and CMV‐LS (subgroup II, represented in white) exchanging untranslated regions (UTRs) and open reading frames (ORFs) in RNA3. Results from both reverse transcriptase‐polymerase chain reaction (RT‐PCR) and visual symptoms of virus are indicated as infected (+) or non‐infected (–). IGR, intergenic region. (b) Photographs of virus symptoms at 20 days post‐inoculation (dpi) and RT‐PCR of upper leaves taken at 20 dpi in melon genotypes Songwhan Charmi (SC) harbouring the cmv1 allele, Piel de Sapo (PS) harbouring the CMV1 allele and SC12‐1‐99 (99) harbouring the cmv1 allele, inoculated with F3(MP1LS) or L3(MP1FNY). C‐, mock‐inoculated plants; M1, Lambda marker (EcoRI, HindIII); pUC, pUC Mix Marker 8 (Fermentas, Waltham, MA, USA).
Using the program SCRATCH protein predictor (http://scratch.proteomics.ics.uci.edu/), which predicts domains based on the predicted secondary structure and relative solvent accessibility for each residue, we observed that it predicted two well‐differentiated domains in the MPs of the two strains: domain 1 from residue 1 to residue 225 and domain 2 from residue 226 to residue 279. A close look at the sequence of both MP genes showed that there was a Bpu1102I restriction site shared by both strains at nucleotide position 628 (209 amino acids). Therefore, the Bpu1102I restriction site separates most of domain 1 (209 of 225 amino acids, the remaining 16 residues being identical or much conserved; see Fig. 4a) from domain 2. Thus, the Bpu1102I site was used to exchange domain 1 of the gene to obtain clones with only the 628 5′‐terminal nucleotides of the FNY MP coding sequence in the background of LS [L3(MP1FNY)] and, likewise, the complementary clone with the first 628 nucleotides of the LS MP coding sequence in the background of FNY [F3(MP1LS)]. After testing the virulence of the resulting clones in the resistant SC12‐1‐99 line, the only one that produced systemic infection was the clone carrying domain 1 of FNY in the background of LS [Fig. 3, clone L3(MP1FNY)]. Therefore, this indicated that the virulence was conferred either by one or several residues present in the N‐terminal 209 amino acids of domain 1 of the MP of FNY, or by the tertiary structure of the protein generated when these residues were present.
Figure 4.
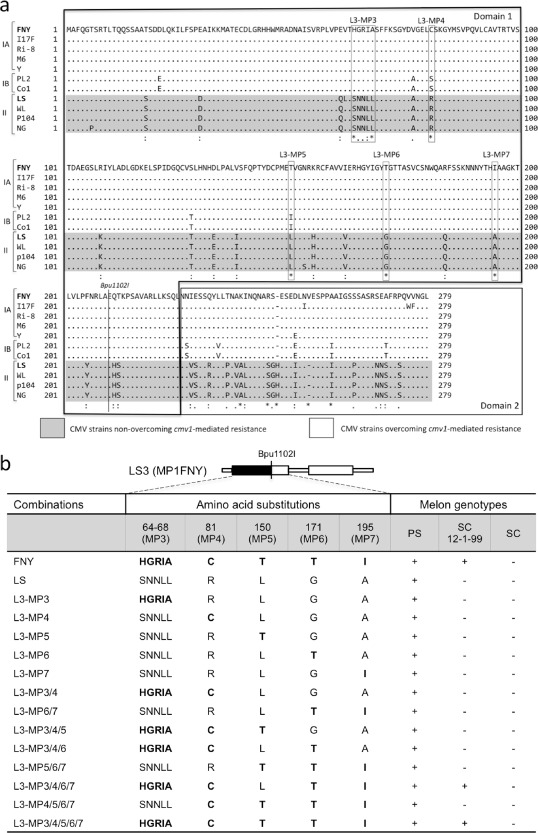
Comparison of the movement protein (MP) of subgroup I and II strains and amino acid substitutions. (a) Alignment between MPs of strains of subgroups I and II showing the two domains, Bpu1102I restriction site and location of amino acid substitutions. CMV‐FNY and CMV‐LS strains are indicated in bold. Non‐conserved amino acids are indicated as ‘*’, semi‐conserved amino acids are indicated as ‘.’ and conserved amino acid substitutions are indicated as ‘:’ below the sequences. (b) Representation and results of single and multiple amino acid substitutions. Amino acids of FNY and LS are indicated in bold and plain lettering, respectively. The presence of virus in the upper leaves was observed by reverse transcriptase‐polymerase chain reaction (RT‐PCR) at 21 days post‐inoculation (dpi) in the melon genotypes Piel de Sapo (PS) harbouring the CMV1 allele, Songwhan Charmi (SC) harbouring the cmv1 allele and SC12‐1‐99 harbouring the cmv1 allele. +, infected plant; –, non‐infected plant.
Identification of the residues relevant for virulence
A comparison of the MPs of the two strains showed that they were 83.5% identical in residues and 93.1% similar. The comparison of the corresponding N‐terminal 209 amino acids of the two strains showed that they were 89.5% identical and differed in 22 amino acids, and that only in five positions were there amino acid changes with very different chemical properties, more likely to produce important changes in the final structure. There was a group of five amino acids that changed as a block at positions 64–68 (SNNLL in LS to HGRIA in FNY), and four unique changes: R81C, L150T, G171T and A195I. A comparison between the MPs of the 11 CMV strains belonging to subgroups I and II used above showed that they were also different in these same five positions, although, inside subgroup I, there were different residues at positions MP4 and MP5 in the strains of subgroup IB than in those of subgroup IA (Fig. 4a). Therefore, the presence of these four residues, and one group of residues in all subgroup I strains, was a general feature for all the strains that overcame the resistance provided by cmv1.
To assign virulence to a particular residue or group of residues, we performed site‐directed mutagenesis in the MP gene of LS to introduce the corresponding FNY residue. First, we introduced the mutations in independent clones to test the effect of each change on virulence, but the inoculation of these mutants into the resistant line 12‐1‐99 did not produce infection (Fig. 4b). Therefore, none of the five mutations individually can produce a gain‐of‐function mutant able to overcome the resistance provided by cmv1. Then, combinations of two, three, four and five changes were generated. Although all were infectious in the susceptible plant PS, inoculation of these clones into SC12‐1‐99 resistant plants revealed that the infectivity of the strain LS was restored when the changes 64–68 (SNNLL to HGRIA), R81C, G171T and A195I were present in the same clone (L3‐MP3/4/6/7, see Fig. 4b), producing infection after 10 dpi. Consistently, the clone L3‐MP3/4/5/6/7, carrying the five mutations, was also infectious. The clones L3‐MP3/4 and L3‐MP6/7 were not infectious in the resistant line, indicating that the changes in the cluster 64–68 (SNNLL to HGRIA), and the point mutations R81C, G171T and A195I, are required changes to overcome cmv1 resistance, and that these are the only four mutations required. The mutation L150T was therefore dispensable for this phenotype. In all cases, the relevant fragment of the progeny viruses was sequenced, confirming that the only mutations present were those introduced for this experiment.
Discussion
We analysed 11 CMV strains for their ability to overcome cmv1‐mediated resistance in the melon accession SC, and established that this resistance was only efficient against strains of subgroup II, whereas strains of subgroup I could infect systemically the resistant line SC12‐1‐99. Subgroups I and II of CMV strains have been defined mainly on the basis of sequences of the CP and RNA3 5′UTR (Roossinck et al., 1999). A phylogeny based on the sequence of MP of the 11 strains used in this work confirms that strains FNY, I17F, Ri‐8, M6 and Y belong to subgroup IA, Pl2 and Co1 belong to subgroup IB, and p104.82, WL, LS and NG belong to subgroup II (Fig. S1, see Supporting Information). The correlation of CMV subgroups with traits has only been observed with regard to temperature and symptoms in tobacco plants (Marchoux et al., 1976; Zhang et al., 1994). Strains of subgroup I are also called heat resistant, as they can infect plants at a higher temperature than can strains of subgroup II, also called heat sensitive (Marchoux et al., 1976). To our knowledge, this is the first resistance phenotype in which the two subgroups show a different response to the resistance gene. Subgroups I and II are distributed worldwide, without any clear prevalence in any geographical area (Palukaitis and Garcia‐Arenal, 2003). However, subgroup IB is more frequent in East Asia. In this study, we observed that the presence of a gene, cmv1, in a Korean melon cultivar conferred resistance against strains of subgroup II only, suggesting that some selection pressure may have occurred. This selection pressure has involved the selection of a cluster of five residues and three additional individual residues in the MP, which may possibly be necessary to maintain a protein structure able to overcome the resistance conferred by cmv1. More work is needed to understand the epidemiological significance of this correlation between cmv1 and resistance to strains of subgroup II. In addition, this observation could allow the development of strategies of breeding to select melon plants resistant to CMV strains of subgroup II.
The ability to invade a plant systemically involves short‐ and long‐distance movement and, in both, viral and host factors are involved. The viral factors are the elements that enable the virus to overcome plant barriers. The analysis of the determinant of virulence that enables CMV to overcome cmv1‐mediated resistance established that the determinant is in the MP coding region. Although the most frequent determinant of virulence in CMV is the CP (Mochizuki and Ohki, 2011), the MP has also been reported to play the main role in virulence in different hosts. In soybean, Hong et al. (2007) constructed chimaeras between two strains (CMV‐SC, which was able to infect the cultivar Hyuongo, and CMV‐SD, which was unable), and reported the MP as a determinant that enabled the CMV‐SC strain to infect this cultivar. In cucurbits, the CP and MP have mostly been implicated in viral spread. The MP has been implicated in slower cell‐to‐cell movement in the inoculated cotyledons and also in systemic spread in squash (Choi et al., 2005; Gal‐On et al., 1996; Kaplan et al., 1997). The CP controls systemic infection in squash (Thompson et al., 2006; Wong et al., 1999) and cucumber (Salánki et al., 2011; Taliansky and García‐Arenal, 1995). Sometimes, the determinant is not a single gene, but a combination of viral gene products. For example, a combination of residues in both the MP and CP determines viral spread in bottle gourd (Takeshita et al., 2001). In the CMV–melon system reported here, we did not detect a role for the CP alone or in combination with the MP in overcoming the resistance provided by cmv1. However, there are at least two other QTLs that contribute, cooperating with cmv1, to the resistance to strains of subgroup I in melon (Guiu‐Aragonés et al., 2014). The CP (and other CMV genes) may also be involved in determining the virulence against one or both of these QTLs. Moreover, CMV is thought to move systemically as particles (Blackman et al., 1998), and therefore it is likely that the CP has a role in the systemic movement of the strains of subgroup I by interacting with the products of these QTLs.
Although CMV does not move cell to cell as virions (Kaplan et al., 1998), the CMV CP is needed for cell‐to‐cell movement, together with MP. The two proteins communicate through a region of 33 C‐terminal amino acids of the MP that determine the dependence or independence of the CP to move the viral genome (Kim et al., 2004; Nagano et al., 2001). In the melon accession SC, the CP of CMV‐LS is able to move the chimaeric virus LS3(MPFNY) that carries the MP of FNY, as this virus produces a full systemic infection. Therefore, the C‐terminal region of the MP does not affect the crosstalk between the two proteins when they belong to different strains, despite being the most divergent part of this protein between both strains (Fig. 4a). Moreover, we have shown that the MP fragment involved in overcoming the resistance of cmv1 is the N‐terminal domain, and therefore interaction with the CP should not be involved in this phenotype. Indeed, this region of the MP has been suggested to be involved in host‐specific functions in both CMV and Brome mosaic virus (Fujita et al., 1996; Hong et al., 2007; Li et al., 2001).
We generated a series of mutant clones in the MP of LS by introducing the corresponding FNY residues, and observed that a combination of four mutations [the group 64–68 (SNNLL to HGRIA) and the point mutations R81C, G171T and A195I] is sufficient to overcome the resistance provided by cmv1. As the structure of the CMV MP has not been resolved, it is difficult to visualize the impact of these mutations in the folding of the protein. However, some functional domains have been reported (Kaplan et al., 1997; Li et al., 2001). There is a hydrophobic core domain between residues 86 to 108, an RNA‐binding domain between amino acids 174 and 233 (Vaquero et al., 1997) and a cysteine‐rich, putative zinc finger domain between amino acids 126 and 194, with two nucleic acid‐binding domains also included in this region (Sasaki et al., 2006). Mutants in these domains have revealed implications in MP functions. In this report, the four changes that together restore the virulence in the cmv1 plant are dispersed in different domains: MP6 and MP7 are in the putative zinc finger domain, MP4 is in the hydrophobic core and the group 64–68 (SNNLL to HGRIA) is located outside of putative domains. MP5, the only change not affecting the gain‐of‐function phenotype, is outside of any predicted domain. The chemical nature of the mutations introduced in LS MP predicts relevant changes in the tertiary structure of the protein. For example, the program SCRATCH protein predictor, mentioned above, predicts three disulfide bonds in FNY MP and two in LS MP, making the structure of LS MP more relaxed. The mutation R81C would recover this disulfide bond and, with it, a more FNY‐like tertiary structure of the MP. FNY MP could possibly interact directly or indirectly with both the PS CMV1 protein and the resistant SC cmv1 protein to continue the infection, whereas LS MP would only interact directly or indirectly with the susceptible PS CMV1 protein to produce a systemic infection. The conformation of the mutant L3‐MP3/4/6/7 would keep the interaction FNY‐like, enabling crosstalk with both the PS and SC alleles of cmv1.
Natural mutations breaking recessive resistances have been found in a few families, and normally they affect the protein that interacts with the resistance gene. Most have been found in members of the Potiviridae family and lie in the viral genome‐linked (VPg) or cylindrical inclusion (CI) genes, which interact directly or indirectly with eukaryotic translation initiation factor (eIF)4E or G, the products of the recessive resistance gene (Moury et al., 2004; Sorel et al., 2014). A single mutation breaking the resistance to cyvv1 and cyvv2 genes (eIF4E) has also been found in the P1 of the potyvirus Clover yellow vein virus, although, in this case, it is not clear how P1 interacts with eIF4E (Nakahara et al., 2010). A single mutant in Rice yellow mottle virus VPg, a member of the genus Sobemovirus, is able to break the resistance against the gene rymv‐1 [eIF4(iso)G] (Hébrard et al., 2006). A resistant‐breaking isolate of Melon necrotic spot carmovirus maps to a few nucleotides in the 3′UTR, the region interacting with eIF4E, the product of the resistance gene nsv (Díaz et al., 2004; Truniger et al., 2008). cmv1 has not been isolated to date. However, it will not be an eIF, as there is no eIF mapping in the cmv1 interval of the melon genome (Essafi et al., 2009). Moreover, we have shown that cmv1‐mediated resistance does not act at the level of translation/replication or short‐distance movement, as CMV‐LS accumulates in the inoculated cotyledons of the resistant line. The implication of the MP as the determinant of virulence in the resistance mediated by cmv1 suggests that it will be related to the impairment of long‐distance movement. Further experiments aimed at addressing the characterization of this resistance will determine whether this is the case.
Experimental Procedures
Plant and virus materials
The genotypes of melon (Cucumis melo L.) used for the study of the resistance to CMV were the Korean accession PI 161375 cultivar ‘Songwhan Charmi’ (SC) and the Spanish cultivar Piel de Sapo, line T111 (PS), as resistant and susceptible controls, respectively. The near‐isogenic line (NIL) SC12‐1‐99 was derived from the NIL SC12‐1 (Essafi et al., 2009), carrying a shorter introgression of SC on the linkage group XII that contains the cmv1 gene. Seeds were pre‐germinated by soaking them in water overnight and then kept for 2–4 days in continuous light at 28 °C. Seedlings were grown in growth chambers (Sanyo MLR‐350H, Sanyo Electric Biomedical Co, Osaka, Japan) in long‐day conditions consisting of 22 °C for 16 h with 5000 lx of light and 18 °C for 8 h in the dark for all infections.
Viruses used in this study were the infectious clones of CMV‐LS (L1, L2 and L3, corresponding to RNAs 1, 2 and 3, respectively) belonging to subgroup II, and CMV‐FNY (F109, F209 and F309, corresponding to RNAs 1, 2 and 3, respectively) belonging to subgroup I (Rizzo and Palukaitis, 1990; Zhang et al., 1994). Other CMV strains used were M6 (Diaz et al., 2003), I17F (Jacquemond and Lauquin, 1988), Ri‐8, Co1, Pl2 (Aramburu et al., 2007) and Y from subgroup I, and NG (Aramburu et al., 2007), WL (Namba et al., 1991) and P104.82 from subgroup II.
Inoculations and virus detection
Viral inocula were freshly prepared from infected zucchini squash Chapin F1 (Semillas Fito SA, Barcelona, Spain) and the sap obtained was rub inoculated onto the cotyledons of 7–10‐day‐old melon plants. For each experiment, 8–10 plants of lines SC12‐1‐99, PS or SC were inoculated. Infectious RNAs of the pseudo‐recombinants and chimaeric viruses were inoculated onto N. benthamiana leaves. RNAs were generated from 1 μg of the linearized infectious cDNA clones by ‘in vitro’ transcription using T7 RNA polymerase (Roche Diagnostics, Mannheim, Germany) and Cap Analog (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's protocols. Infectious clones from CMV‐FNY (Rizzo and Palukaitis, 1990), chimaeric viruses and amino acid substitution mutant clones were linearized using PstI. Infectious clones CMV‐L1, CMV‐L2 and CMV‐L3 were linearized with NotI, HindIII and PstI, respectively (Zhang et al., 1994). The three transcribed RNAs, without further purification, were rub inoculated together onto N. benthamiana leaves. Sap produced from systemically infected leaves was used to inoculate melon cotyledons. Symptoms in melon plants were scored visually 20 days after inoculation. Viral detection was performed by reverse transcriptase‐polymerase chain reaction (RT‐PCR) from young newly developed leaves. For RT‐PCR, RNA was isolated using TriReagent (Sigma‐Aldrich, St Louis, MO, USA) following the manufacturer's protocol. RT‐PCR was performed using PrimeScript (Takara Biotechnology, Dalian, China) and Taq polymerase (Promega Corporation, Madison, WI, USA) according to the manufacturer's instructions, as reported previously (Essafi et al., 2009). Primers used for RT and PCR are listed in Table S1 (see Supporting Information). Combinations of primers from both strains, CMV‐LS and CMV‐FNY, were used to confirm viral infection, when necessary. To confirm the amino acid substitution mutants, the PCR products were sequenced using primer LS3‐1F (Table S1).
Northern blot analysis was performed following Díaz et al. (2004). Briefly, 6 μg of total RNA obtained from inoculated cotyledons were denatured, electrophoresed in 1% agarose gel and vacuum transferred (VacuGene XL, HealthCare, Friburg, Germany) onto a positively charged nylon membrane (Roche Diagnostics, Barcelona, Spain). Samples were crosslinked under UV light (Crosslinker RPN 2500, Amersham Life Science, Friburg, Germany). For hybridizations, a digoxygenin‐11‐UTP‐labelled RNA probe was synthesized from a construct (p73) containing partial sequences of CMV‐LS and CMV‐FNY CP genes using a digoxigenin‐labelling and detection kit (Roche Diagnostics, Barcelona, Spain). For transcription, the plasmid was linearized with ApaI, and cRNA was synthesized using SP6 RNA polymerase (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, UK). Hybridization was performed at 65 °C. Washing steps and detection with the alkaline phosphatase chemiluminescent substrate CDP‐Star (Roche Diagnostics, Barcelona, Spain) were performed as recommended by the manufacturer.
Construction of chimaeric viruses and amino acid substitution mutants
Chimaeric viruses between CMV‐LS and CMV‐FNY were generated by exchanging all the UTRs and ORFs separately. Briefly, overlapping PCR fragments sharing around 20 nucleotides at the ends were amplified. For each pair of fragments, the overlapping sequence contained part of LS and part of FNY. Gel‐purified PCR fragments were submitted to chimaeric PCR performed in two steps. The first eight cycles were based on the temperature given by the overlapping sequence, usually higher than the temperature of the remaining cycles, given by the flanking primers. These cycles amplified the chimaeric PCR generated during the first cycles. The chimaeric fragments were exchanged in the viral construct using unique restriction sites. In the cases of ORF exchanges and IGR exchanges, three PCR fragments were amplified, creating two overlapping regions, and then the chimaeric PCR assembled the three fragments. All PCRs were performed in the presence of 0.2 mm deoxynucleotide triphosphates (dNTPs) with Pfu DNA polymerase (Promega Corporation) and specific primers (Table S2).
Constructs in which the 209 N‐terminal amino acids of the MP were exchanged were generated using restriction sites. In the case of the construct F3(MP1LS), the chimaeric construct L3(5′UTRFNY) was digested with Bpu1102I and XmaI, and the fragment was cloned into the same restriction sites of the construct F309, which carries the whole RNA3 of FNY, to have only the 209 amino acids of LS in the background of FNY. The complementary construct L3 (MP1FNY) was performed using the same approach on the clone F3(5′UTRLS), and cloned using the same restriction sites on L3. The amino acid substitution mutants introducing specific residues of MP‐FNY into MP‐LS were built using a GENEART® Site‐Directed Mutagenesis System (Invitrogen Life Technologies, Carlsbad, CA, USA), following the manufacturer's instructions; 20 ng of DNA of plasmid LS3 were hybridized to the mutagenic primer and allowed to polymerize in the presence of 0.2 mm dNTPs and Pfu DNA polymerase (Promega Corporation). All mutagenic primers used are listed in Table S3 (see Supporting Information). The substitutions were performed independently at five positions. Multiple amino acid combinations were performed using the same technique over constructs carrying previously introduced mutations. Sequences to confirm all chimaeric constructs were analysed using Sequencher™ version 4.8 (Gene Codes Corporation; Ann Arbor, MI, USA).
MP sequences and alignment
Sequences of the MP genes of all the strains used were obtained by performing RNA extraction and RT‐PCR of infected material, as described previously (Essafi et al., 2009). The coding sequence of MPs of strains of subgroup II were sequenced with primers specific for LS (LS3‐1F, 5′GTAATCTTACCACTTTCTTTTC; LS3‐400F, 5′GTTTCTACGGATGCTGAGG) and LS3‐1400R (5′CTTTCACTACCCACGAAGG). Primers used for strains of subgroup I were specific for CMV‐FNY (F309‐1F, 5′GTAATCTTACCACTGTGTGTG; F309‐400F, 5′TACTCGAACAGTTTCCACTG) and F309‐1400R (5′GTTAATAGTTGGACGACCAG). Alignment of MPs was performed using Protein‐Protein BLAST 2.2.25. The sequences of the MP of strains FNY, I17F, Ri‐8, Y and LS were identical to the corresponding sequences published in GenBank: FNY, BAA01396.1; I17F, CAA77064.1; Ri‐8, CAJ65583.1; Y, AAA46419.1; LS, AAD45246.1.
Accession numbers for the new sequences
Accession numbers for the sequences of strains M6, P104.82, WL, Pl2, Co1 and NG in GenBank are KM114893, KM114894, KM114895, KM114896, KM114897 and KM114898, respectively.
Supporting information
Fig. S1 Phylogenetic tree of the 11 Cucumber mosaic virus (CMV) strains. FNY, I17F, Ri‐8, M6 and Y belong to subgroup IA, Pl2 and Co1 belong to subgroup IB, and p104.82, WL, LS and NG belong to subgroup II.
Table S1 Specific combinations of primers used for virus detection in plants inoculated with pseudo‐recombinants, chimaeras with untranslated region (UTR) and open reading frame (ORF) exchanges and amino acid substitution mutants.
Table S2 Primers used to generate untranslated region (UTR) and open reading frame (ORF) exchanged regions.
Table S3 Primers used for site‐directed mutagenesis.
Acknowledgements
We thank Fuensanta Garcia for technical support, J. J. López‐Moya and Jordi Garcia‐Mas for critical reading, Marc Tormo for helping with the protein alignments, P. Palukaitis for CMV‐FNY and LS clones, M. Luis‐Arteaga for CMV P104.82, E. Moriones for CMV M6, J. Aramburu for CMV Ri‐8, I17F, Co‐1, Pl‐2 and NG, and F. Garcia‐Arenal for CMV Y. This work was supported by grants AGL2009‐12698‐C02‐01 and AGL2012‐40130‐C02‐01 from the Spanish Ministry of Science and Innovation. C.G‐A was supported by grant BES‐2010‐030274 from the Spanish Ministry of Science and Innovation.
We declare that we have no conflict of interest.
References
- Albiach‐Marti, M. , Robertson, C. , Gowda, S. , Tatineni, S. , Belliure, B. , Garnsey, S. , Folimonova, S. , Moreno, P. and Dawson, W. (2010) The pathogenicity determinant of Citrus tristeza virus causing the seedling yellows syndrome maps at the 3′‐terminal region of the viral genome. Mol. Plant Pathol. 11, 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramburu, J. , Galipienso, L. and López, C. (2007) Reappearance of Cucumber mosaic virus isolates belonging to subgroup IB in tomato plants in north‐eastern Spain. J. Phytopathol. 155, 513–518. [Google Scholar]
- Blackman, L.M. , Boevink, P. , Santa Cruz, S. , Palukaitis, P. and Oparka, K.J. (1998) The movement protein of Cucumber mosaic virus traffics into sieve elements in minor veins of Nicotiana clevelandii . Plant Cell, 10, 525–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, S. , Palukaitis, P. , Min, B. , Lee, M. , Choi, J. and Ryu, K. (2005) Cucumber mosaic virus 2a polymerase and 3a movement proteins independently affect both virus movement and the timing of symptom development in zucchini squash. J. Gen. Virol. 86, 1213–1222. [DOI] [PubMed] [Google Scholar]
- Choi, S.H. , Hagiwara‐Komoda, Y. , Nakahara, K.S. , Atsumi, G. , Shimada, R. , Hisa, Y. et al (2013) Quantitative and qualitative involvement of P3N‐PIPO in overcoming recessive resistance against Clover yellow vein virus in pea carrying the cyv1 gene. J. Virol. 87, 7326–7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroocq, V. , Salvador, B. , Sicard, O. , Glasa, M. , Cosson, P. , Svanella‐Dumas, L. et al (2009) The determinant of potyvirus ability to overcome the RTM resistance of Arabidopsis thaliana maps to the N‐terminal region of the coat protein. Mol. Plant–Microbe Interact. 22, 1302–1311. [DOI] [PubMed] [Google Scholar]
- Diaz, J.A. , Mallor, C. , Soria, C. , Camero, R. , Garzo, E. , Fereres, A. et al (2003) Potential sources of resistance for melon to nonpersistently aphid‐borne viruses. Plant Dis. 87, 960–964. [DOI] [PubMed] [Google Scholar]
- Díaz, J. , Nieto, C. , Moriones, E. , Truniger, V. and Aranda, M. (2004) Molecular characterization of a Melon necrotic spot virus strain that overcomes the resistance in melon and nonhost plants. Mol. Plant–Microbe Interact. 17, 668–675. [DOI] [PubMed] [Google Scholar]
- Diaz‐Pendon, J. , Li, F. , Li, W.‐X. and Ding, S.‐W. (2007) Suppression of antiviral silencing by Cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell, 19, 2053–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divéki, Z. , Salánki, K. and Balázs, E. (2004) The necrotic pathotype of the Cucumber mosaic virus (CMV) Ns strain is solely determined by amino acid 461 of the 1a protein. Mol. Plant–Microbe Interact. 17, 837–845. [DOI] [PubMed] [Google Scholar]
- Edwardson, J.R. and Christie, R.G. (1991) Cucumoviruses In: CRC Handbook of Viruses Infecting Legumes (Edwardson J.R. and Christie R.G., eds), pp. 293–319. Boca Raton, FL: CRC Press. [Google Scholar]
- Essafi, A. , Diaz‐Pendon, J.A. , Moriones, E. , Monforte, A.J. , Garcia‐Mas, J. and Martin‐Hernandez, A.M. (2009) Dissection of the oligogenic resistance to Cucumber mosaic virus in the melon accession PI 161375. Theor. Appl. Genet. 118, 275–284. [DOI] [PubMed] [Google Scholar]
- Fujita, Y. , Mise, K. , Okuno, T. , Ahlquist, P. and Furusawa, I. (1996) A single codon change in a conserved motif of a bromovirus movement protein gene confers compatibility with a new host. Virology, 223, 283–291. [DOI] [PubMed] [Google Scholar]
- Gal‐On, A. , Kaplan, I.B. and Palukaitis, P. (1996) Characterization of Cucumber mosaic virus: II. Identification of movement protein sequences that influence its accumulation and systemic infection in tobacco. Virology, 226, 354–361. [DOI] [PubMed] [Google Scholar]
- Guiu‐Aragonés, C. , Monforte, A.J. , Saladié, M. , Corrêa, R.X. , Garcia‐Mas, J. and Martín‐Hernández, A.M. (2014) The complex resistance to Cucumber mosaic cucumovirus (CMV) in the melon accession PI 161375 is governed by one gene and at least two quantitative trait loci. Mol. Breed. 34, 351–362. [Google Scholar]
- Hébrard, E. , Pinel‐Galzi, A. , Bersoult, A. , Siré, C. and Fargette, D. (2006) Emergence of a resistance‐breaking isolate of Rice yellow mottle virus during serial inoculations is due to a single substitution in the genome‐linked viral protein VPg. J. Gen. Virol. 87, 1369–1373. [DOI] [PubMed] [Google Scholar]
- Hong, J.S. , Ohnishi, S. , Masuta, C. , Choi, J.K. and Ryu, K.H. (2007) Infection of soybean by Cucumber mosaic virus as determined by viral movement protein. Arch. Virol. 152, 321–328. [DOI] [PubMed] [Google Scholar]
- Jacquemond, M. and Lauquin, G. (1988) The cDNA of Cucumber mosaic virus‐associated satellite RNA has in vivo biological properties. Biochem. Biophys. Res. Commun. 151, 388–395. [DOI] [PubMed] [Google Scholar]
- Kang, W.‐H. , Seo, J.‐K. , Chung, B. , Kim, K.‐H. and Kang, B.‐C. (2012) Helicase domain encoded by Cucumber mosaic virus RNA1 determines systemic infection of Cmr1 in pepper. PLoS ONE, 7, e43136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, I. , Gal‐On, A. and Palukaitis, P. (1997) Characterization of Cucumber mosaic virus. III. Localization of sequences in the movement protein controlling systemic infection in cucurbits. Virology, 230, 343–349. [DOI] [PubMed] [Google Scholar]
- Kaplan, I. , Zhang, L. and Palukaitis, P. (1998) Characterization of Cucumber mosaic virus. V. Cell‐to‐cell movement requires capsid protein but not virions. Virology, 246, 221–231. [DOI] [PubMed] [Google Scholar]
- Karchi, Z. , Cohen, S. and Govers, A. (1975) Inheritance of resistance to Cucumber mosaic virus in melons. Phytopathology, 65, 479–481. [Google Scholar]
- Kim, C. and Palukaitis, P. (1997) The plant defense response to Cucumber mosaic virus in cowpea is elicited by the viral polymerase gene and affects virus accumulation in single cells. EMBO J. 16, 4060–4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M.J. , Huh, S.U. , Ham, B.‐K. and Paek, K.‐H. (2008) A novel methyltransferase methylates Cucumber mosaic virus 1a protein and promotes systemic spread. J. Virol. 82, 4823–4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.H. , Kalinina, N.O. , Andreev, I. , Ryabov, E.V. , Fitzgerald, A.G. , Taliansky, M.E. et al (2004) The C‐terminal 33 amino acids of the Cucumber mosaic virus 3a protein affect virus movement, RNA binding and inhibition of infection and translation. J. Gen. Virol. 85, 221–230. [DOI] [PubMed] [Google Scholar]
- Lewsey, M. , Robertson, F.C. , Canto, T. , Palukaitis, P. and Carr, J.P. (2007) Selective targeting of miRNA‐regulated plant development by a viral counter‐silencing protein. Plant J. 50, 240–252. [DOI] [PubMed] [Google Scholar]
- Li, Q. , Ryu, K. and Palukaitis, P. (2001) Cucumber mosaic virus–plant interactions: identification of 3a protein sequences affecting infectivity, cell‐to‐cell movement, and long‐distance movement. Mol. Plant–Microbe Interact. 14, 378–385. [DOI] [PubMed] [Google Scholar]
- Mansilla, C. , Sánchez, F. , Padgett, H. , Pogue, G. and Ponz, F. (2009) Chimeras between Oilseed rape mosaic virus and Tobacco mosaic virus highlight the relevant role of the tobamoviral RdRp as pathogenicity determinant in several hosts. Mol. Plant Pathol. 10, 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchoux, G. , Douine, L. and Quit, J.B. (1976) Comportement thermique differential de certaines souches du virus de la mosaique du concombre. Hypothése d'un mecanism pleiotropique reliant plusieurs proprietés. C. R. Acad. Sci. Paris Ser. D, 283, 1601–1604. [PubMed] [Google Scholar]
- Mochizuki, T. and Ohki, S. (2011) Cucumber mosaic virus: viral genes as virulence determinants. Mol. Plant Pathol. 13, 217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moury, B. , Morel, C. , Johansen, E. , Guilbaud, L. , Souche, S. , Ayme, V. et al (2004) Mutations in potato virus Y genome‐linked protein determine virulence toward recessive resistances in Capsicum annuum and Lycopersicon hirsutum . Mol. Plant–Microbe Interact. 17, 322–329. [DOI] [PubMed] [Google Scholar]
- Nagano, H. , Mise, K. , Furusawa, I. and Okuno, T. (2001) Conversion in the requirement of coat protein in cell‐to‐cell movement mediated by the Cucumber mosaic virus movement protein. J. Virol. 75, 8045–8053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara, K.S. , Shimada, R. , Choi, S.‐H. , Yamamoto, H. , Shao, J. and Uyeda, I. (2010) Involvement of the P1 cistron in overcoming eIF4E‐mediated recessive resistance against Clover yellow vein virus in pea. Mol. Plant–Microbe Interact. 23, 1460–1469. [DOI] [PubMed] [Google Scholar]
- Namba, S. , Ling, K.S. , Gonsalves, C. , Gonsalves, D. and Slightom, J.L. (1991) Expression of the gene encoding the coat protein of Cucumber mosaic virus (CMV) strain WL appears to provide protection to tobacco plants against infection by several different CMV strains. Gene, 107, 181–188. [DOI] [PubMed] [Google Scholar]
- Palukaitis, P. and Garcia‐Arenal, F. (2003) Cucumoviruses. Adv. Virus Res. 62, 241–323. [DOI] [PubMed] [Google Scholar]
- Rizzo, T.M. and Palukaitis, P. (1990) Construction of full‐length cDNA clones of cucumber mosaic virus RNAs 1, 2 and 3: generation of infectious RNA transcripts. Mol. Gen. Genet. 222, 249–256. [DOI] [PubMed] [Google Scholar]
- Roossinck, M.J. (2001) Cucumber mosaic virus, a model for RNA virus evolution. Mol. Plant Pathol. 2, 59–63. [DOI] [PubMed] [Google Scholar]
- Roossinck, M.J. , Zhang, L. and Hellwald, K.‐H. (1999) Rearrangements in the 5′ nontranslated region and phylogenetic analyses of Cucumber mosaic virus RNA3 indicate radial evolution of three subgroups. J. Virol. 73, 6752–6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryabov, E.V. , Roberts, I.M. , Palukaitis, P. and Taliansky, M. (1999) Host‐specific cell‐to‐cell and long‐distance movements of Cucumber mosaic virus are facilitated by the movement protein of groundnut rosette virus. Virology, 260, 98–108. [DOI] [PubMed] [Google Scholar]
- Ryu, K.H. , Kim, C.‐H. and Palukaitis, P. (1998) The coat protein of Cucumber mosaic virus is a host range determinant for infection of maize. Mol. Plant–Microbe Interact. 11, 351–357. [Google Scholar]
- Salánki, K. , Kiss, L. , Gellért, A. and Balázs, E. (2011) Identification of a coat protein region of Cucumber mosaic virus (CMV) essential for long‐distance movement in cucumber. Arch. Virol. 156, 2279–2283. [DOI] [PubMed] [Google Scholar]
- Sasaki, N. , Park, J.‐W. , Maule, A. and Nelson, R. (2006) The cysteine–histidine‐rich region of the movement protein of Cucumber mosaic virus contributes to plasmodesmal targeting, zinc binding and pathogenesis. Virology, 349, 396–408. [DOI] [PubMed] [Google Scholar]
- Sorel, M. , Svanella‐Dumas, L. , Candresse, T. , Acelin, G. , Pitarch, A. , Houvenaghel, M.C. et al (2014) Key mutations in the cylindrical inclusion involved in Lettuce mosaic virus adaptation to eIF4E‐mediated resistance in lettuce. Mol. Plant–Microbe Interact. 27, 1014–1024. [DOI] [PubMed] [Google Scholar]
- Takahashi, H. , Suzuki, M. , Natsuaki, K. , Shigyo, T. , Hino, K. , Teraoka, T. et al (2001) Mapping the virus and host genes involved in the resistance response in Cucumber mosaic virus‐infected Arabidopsis thaliana . Plant Cell Physiol. 42, 340–347. [DOI] [PubMed] [Google Scholar]
- Takeshita, M. , Suzuki, M. and Takahami, Y. (2001) Combination of amino acids in the 3a protein and the coat protein of Cucumber mosaic virus determines symptom expression and viral spread in bottle gourd. Arch. Virol. 146, 697–711. [DOI] [PubMed] [Google Scholar]
- Taliansky, M. and García‐Arenal, F. (1995) Role of cucumovirus capsid protein in long‐distance movement within the infected plant. J. Virol. 69, 916–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.R. , Doun, S. and Perry, K.L. (2006) Compensatory capsid protein mutations in Cucumber mosaic virus confer systemic infectivity in squash (Cucurbita pepo). J. Virol. 80, 7740–7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truniger, V. , Nieto, C. , González‐Ibeas, D. and Aranda, M. (2008) Mechanism of plant eIF4E‐mediated resistance against a Carmovirus (Tombusviridae): cap‐independent translation of a viral RNA controlled in cis by an (a)virulence determinant. Plant J. 56, 716–727. [DOI] [PubMed] [Google Scholar]
- Vaquero, C. , Liao, Y. and Fischer, R. (1997) Mapping of the RNA‐binding domain of the Cucumber mosaic virus movement protein. J. Gen. Virol. 78, 2095–2099. [DOI] [PubMed] [Google Scholar]
- Wong, S.‐M. , Thio, S.S.‐C. , Shintaku, M.H. and Palukaitis, P. (1999) The rate of cell‐to‐cell movement in squash of Cucumber mosaic virus is affected by sequences of the capsid protein. Mol. Plant–Microbe Interact. 12, 628–632. [Google Scholar]
- Zhang, L. , Handa, K. and Palukaitis, P. (1994) Mapping local and systemic symptom determinants of Cucumber mosaic cucumovirus in tobacco. J. Gen. Virol. 75, 3185–3191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Phylogenetic tree of the 11 Cucumber mosaic virus (CMV) strains. FNY, I17F, Ri‐8, M6 and Y belong to subgroup IA, Pl2 and Co1 belong to subgroup IB, and p104.82, WL, LS and NG belong to subgroup II.
Table S1 Specific combinations of primers used for virus detection in plants inoculated with pseudo‐recombinants, chimaeras with untranslated region (UTR) and open reading frame (ORF) exchanges and amino acid substitution mutants.
Table S2 Primers used to generate untranslated region (UTR) and open reading frame (ORF) exchanged regions.
Table S3 Primers used for site‐directed mutagenesis.


