Summary
The AvrE superfamily of type III effectors (T3Es) is widespread among type III‐dependent phytobacteria and plays a crucial role during bacterial pathogenesis. Members of the AvrE superfamily are vertically inherited core effectors, indicating an ancestral acquisition of these effectors in bacterial plant pathogens. AvrE‐T3Es contribute significantly to virulence by suppressing pathogen‐associated molecular pattern (PAMP)‐triggered immunity. They inhibit salicylic acid‐mediated plant defences, interfere with vesicular trafficking and promote bacterial growth in planta. AvrE‐T3Es elicit cell death in both host and non‐host plants independent of any known plant resistance protein, suggesting an original interaction with the plant immune system. Recent studies in yeast have indicated that they activate protein phosphatase 2A and inhibit serine palmitoyl transferase, the first enzyme of the sphingolipid biosynthesis pathway. In this review, we describe the current picture that has emerged from studies of the different members of this fascinating large family.
Keywords: AvrE, PAMP‐triggered immunity, protein phosphatase 2A, receptor‐like kinase, sphingolipid
Introduction
To induce disease in their respective plants, many Gram‐negative plant bacterial pathogens depend on a functional type III secretion system (T3SS), which allows the injection of bacterial proteins, named type III effectors (T3Es), inside plant host cells.
Collectively, the T3E repertoire modulates cellular processes and suppresses host defences for the benefit of the pathogen (Block and Alfano, 2011). Plants, however, have developed effective immune defences to resist pathogen attack. The plant immune system recognizes pathogen‐associated molecular patterns (PAMPs) of the invading pathogen, and triggers PAMP‐triggered immunity (PTI). Bacterial pathogens inject T3Es to counteract and suppress PTI, but plant resistance (R) proteins can sometimes recognize some of the injected T3Es and this triggers a second line of defence, called effector‐triggered immunity (ETI). To counteract ETI, pathogens either lose the recognized T3Es or acquire new T3Es to suppress ETI (Chisholm et al., 2006; Jones and Dangl, 2006; Lindeberg et al., 2012). As a result of this endless arms race, the repertoire of T3Es has been shaped by evolution, is incredibly diverse and varies greatly among bacteria. The AvrE family of T3Es is the only family of TE3s present in all type III‐dependent, agriculturally important phytobacteria, including gammaproteobacteria (enterobacteria, xanthomonads and pseudomonads) and betaproteobacteria (Ralstonia spp.). In this review, we describe the current picture that has emerged from studies of the different members of this large family.
Historical Discovery of the AvrE Family of T3Es
AvrE was originally cloned from Pseudomonas syringae pv. tomato (Pto) PT23 on the basis of its ability to confer avirulence to P. syringae pv. glycinea race 4 in soybean (Kobayashi et al., 1989). A blast‐p search revealed that AvrE shared 28% identity with the DspA/E protein required for Erwinia amylovora pathogenicity, and cross‐complementation analysis confirmed conservation of function (Bogdanove et al., 1998a; Gaudriault et al., 1997). Both encoding genes are located next to the T3SS cluster, and orthologues, also linked to the T3SS cluster, were later described in other plant‐pathogenic bacteria: Pantoea stewartii ssp. stewartii (WtsE) (Frederick et al., 2001), Pantoea agglomerans pv. gypsophilae (DspE/A) (Mor et al., 2001), Dickeya dadantii (DspE) (Glasner et al., 2011) and Pectobacterium atrosepticum and carotovorum (DspE) (Holeva et al., 2004; Kim et al., 2011). Combinatorial deletion analysis of Pto DC3000 T3Es identified HopR1 as a distant AvrE orthologue sharing 14% identity with AvrE. blast‐p search with HopR1 from Pto DC3000 generates hits in Xanthomonas spp. (XopAM) and Ralstonia solanacearum (PopS, also named RipR) (Kvitko et al., 2009; Peeters et al., 2013). The genes encoding HopR1, XopAM and PopS are not located next to the T3SS cluster.
Phylogenetic analysis indicates that AvrE (pseudomonads)/DspA/E, DspE or WtsE (enterobacteria)/PopS (Ralstonia spp.)/HopR (pseudomonads)/XopAM (xanthomonads) sequences can be grouped into five different clades (Jacobs et al., 2013). The phylogenetic trees obtained with PopS or AvrE mirror the phylogenies of Ralstonia and Pseudomonas species, respectively, indicating that the genes were vertically inherited (Jacobs et al., 2013; Rohmer et al., 2004). This highlights the ancestral and crucial role played by this T3E family in plant bacterial pathogenesis.
Contribution to Virulence
Members of the AvrE superfamily contribute significantly to virulence on host plants. DspA/E, WtsE, DspE/A and DspE are essential for the pathogenicity of E. amylovora, Pa. stewartii ssp. stewartii, Pa. agglomerans pv. gypsophilae and Pe. carotovorum, respectively (Frederick et al., 2001; Gaudriault et al., 1997; Kim et al., 2011; Mor et al., 2001). The crucial role played by these effectors could probably be explained by the small repertoire of T3Es found in the genome of these pathogens. However, this result is surprising for the gall‐forming pathogen Pa. agglomerans pv. gypsophilae and the soft rot pathogen Pe. carotovorum, and suggests that disease depends on both WtsE and DspE injection inside the host cell and on the production of phytohormones and plant cell wall‐degrading enzymes (Barash and Manulis‐Sasson, 2007; Kim et al., 2011). Such a dependence was, however, not observed with DspE of Pe. atrosepticum, as DspE makes only a minor contribution to the virulence of this soft rot pathogen on potato stems and tubers (Holeva et al., 2004). Disruption of R. solanacearum PopS, Xanthomonas spp. XopAM or P. syringae AvrE leads to slightly reduced virulence of the mutant strains (Badel et al., 2006; Jacobs et al., 2013; Jiang et al., 2009; Kvitko et al., 2009; Qian et al., 2005). The subtle phenotype observed could be explained by functional overlap among effectors as a result of the large T3E repertoire of the latter bacterial strains. For example, HopR1 and AvrE are part of the same redundant effector group (REG) and both partially complement the ΔIV‐ΔCEL Pto DC3000 mutant, multi‐deleted of several T3Es, including HopR1 and AvrE, for growth on Nicotiana benthamiana (Cunnac et al., 2011; Kvitko et al., 2009). Interestingly, HopR1 does not complement the ΔIV‐ΔCEL Pto DC3000 mutant for growth on tomato, suggesting that the importance of HopR1 may differ in different plants (Kvitko et al., 2009).
Secretion, Translocation and Chaperone Requirement
The secretion of effectors of the AvrE family can be observed in vitro when bacteria are cultivated in minimal medium (Bogdanove et al., 1998b; Gaudriault et al., 1997; Ham et al., 2006). As observed for most T3Es studied to date, N‐terminal uncleaved secretion and translocation signals are described in DspA/E and AvrE (Badel et al., 2006; Bocsanczy et al., 2008; Ham et al., 2009; Oh et al., 2010; Triplett et al., 2009). Genes encoding DspA/E, DspE, AvrE and WtsE are associated with a type III secretion chaperone (T3SC) gene. These T3SCs are required for full accumulation of Pa. stewartii WtsE or E. amylovora DspA/E inside the bacterial cytoplasm. Mutation of the T3SC gene almost abolishes the secretion of E. amylovora DspA/E, but has only a slight impact on Pa. stewartii WtsE secretion, indicating that additional T3SCs are probably involved in WtsE secretion (Gaudriault et al., 2002; Ham et al., 2006). The T3SC of P. syringae AvrE has not been studied in detail; however, mutation in the transcriptional unit that encodes this T3SC abolishes the ability of AvrE to induce a hypersensitive reaction (HR) on soybean cultivar, suggesting that this T3SC is important for AvrE secretion (Lorang and Keen, 1995). Erwinia amylovora DspA/E interaction with its T3SC has been studied in detail. Two independent groups identified a chaperone‐binding domain (CBD) within amino acids 51–200 of DspA/E (Oh et al., 2010; Triplett et al., 2009). Neither disruption of this N‐terminal DspA/E CBD nor deletion of the T3SC gene has a significant impact on translocation levels of N‐terminal DspA/E–CyaA fusions (Triplett et al., 2009), but the presence of T3SC strongly enhances the translocation of a DspA/E–AvrRpt2 fusion protein into plant cells (Oh et al., 2010). These contradictory results indicate that the requirement for the chaperone is influenced by the C‐terminal part of the protein. In accordance with this, it was found that the T3SC also interacts with the C‐terminal half of DspA/E (Triplett et al., 2010). Furthermore, another T3SC has also been found to interact with the C‐terminal domain of DspA/E (Oh et al., 2010). The interaction with several T3SCs and the presence of several CBDs may be required because effectors of the AvrE family are very large proteins.
T3SC genes are not found next to genes encoding HopR1, PopS or XopAM. Whether these effectors do not need chaperone assistance or whether T3SCs encoded elsewhere in the genome assist the secretion/injection of these effectors remains to be determined.
Regulation of Expression
The different genes of the AvrE family are co‐regulated with the T3SS (Gaudriault et al., 1997; Hogan et al., 2013; Jacobs et al., 2013; Lorang and Keen, 1995; McNally et al., 2012; Mor et al., 2001; Yang et al., 2004, 2010). In vitro, their expression is repressed in rich medium and induced in minimal medium (Gaudriault et al., 1997; Peng et al., 2006; Wei et al., 1992). In planta, their expression is detected as early as 2 h following inoculation and is still observed 2–3 days post‐infection, indicating that AvrE‐like effectors are probably required at both the beginning and throughout the infection process (Hogan et al., 2013; Kim et al., 2011; Peng et al., 2006; Pester et al., 2012; Yang et al., 2004). Interestingly, Peng et al. (2006), by analysing the expression of green fluorescent protein (GFP) fused to the dspE promoter of D. dadantii with a fluorescence‐activated cell sorter, observed that a proportion of D. dadantii cells never expressed dspE in various media or host plants. This raises the possibility that individual bacterial cells may play different roles and work together as a community effort to ensure disease development. Wang et al. (2010) also noticed that the expression level of E. amylovora dspA/E correlates with the aggressiveness of the strain, suggesting that this effector induces a dose‐dependent effect.
Physiological Effects of AvrE‐Like Effectors: Suppression of Defence Associated with Perturbation of Cellular Traffic and Slow Death Induction
Several members of the AvrE family are able to suppress salicylic acid‐dependent defence responses, such as callose deposition or PR1 expression (Boureau et al., 2006, 2011; DebRoy et al., 2004; Ham et al., 2008; Jacobs et al., 2013). In contradiction with this general scheme, DspE of Pe. carotovorum is unable to suppress callose deposition induced by Pe. carotovorum along the leaf vein of N. benthamiana (Kim et al., 2011). Whether this reflects intrinsic differences among AvrE effectors, or whether the callose deposition induced by Pe. carotovorum is different from the callose deposition induced by P. syringae, remains to be determined. In support of the second hypothesis, it is known that callose deposition in Arabidopsis thaliana involves more than one pathway (Luna et al., 2011). Furthermore, DspA/E of E. amylovora suppresses P. syringae‐induced, but not E. amylovora‐induced, callose deposition on A. thaliana, further indicating that callose deposition induced by different pathogens is somehow different (Degrave et al., 2013).
When ectopically expressed, most effectors of the AvrE family elicit cell death on host and non‐host plants (Badel et al., 2006; Boureau et al., 2006; Ham et al., 2006, 2008; Hogan et al., 2013; Oh et al., 2007). Whether the cell death observed following expression on non‐host plants is inherent to the toxicity of this effector family, or whether effectors of the AvrE family trigger an HR on non‐host plants as a result of recognition by an R protein, has been a matter of debate. First, one should keep in mind that the answer could vary between non‐host plants, but nobody has yet identified R proteins involved in such recognition. For example, although AvrE was discovered on the basis of its ability to confer avirulence to P. syringae pv. glycinea race 4 in soybean (Kobayashi et al., 1989), it was later found to confer such avirulence to all soybean cultivars tested, raising the possibility that the phenotype observed is simply a result of AvrE toxicity on soybean (Lorang and Keen, 1995). Against recognition by an R protein and ETI triggering, it should be noted that the induced cell death develops slowly (approximately 36 h) on both host and non‐host plants (Badel et al., 2006; Boureau et al., 2006). This ‘slow death’ phenotype is also observed with an A. thaliana transgenic line expressing E. amylovora DspA/E under an oestradiol‐inducible promoter (Degrave et al., 2013). Oestradiol induction allows an investigation of the dose‐dependent effect. When the expression of DspA/E is induced by low oestradiol concentrations, transgenic plants suffer growth reduction without associated cell death. With high oestradiol concentrations, which lead to stronger DspA/E expression, leaves start to wilt at 24 h post‐induction and are completely necrotic at 5 days post‐induction (Degrave et al., 2013). Most surprisingly, transient expression of an R. solanacearum 35S promoter–PopS–haemagglutinin tag (HA) construct does not induce cell death, suggesting that this particular member of the AvrE family could have evolved to avoid cell death induction (Jacobs et al., 2013). This is consistent with the fact that R. solanacearum causes a non‐necrotic wilt. However, one may also question whether the addition of an HA tag at the end of the protein is or is not deleterious for PopS function. Indeed, deletion of the four penultimate amino acids of Pa. stewartii WtsE or E. amylovora DspA/E inactivates the cell death‐inducing ability of these effectors (Ham et al., 2008; A. Degrave et al., unpublished observations).
The expression of DspA/E in transgenic A. thaliana lines is also associated with the inhibition of protein synthesis (Degrave et al., 2013). This is consistent with the fact that Pa. stewartii WtsE‐induced symptoms in corn seedlings are not inhibited by cycloheximide, which blocks de novo protein synthesis in eukaryotic cells (Ham et al., 2006), and the observation that cycloheximide partially rescues the growth of an E. amylovora dspA/E mutant on A. thaliana (Degrave et al., 2013).
AvrE Type III Family Effectors, Large Enigmatic Proteins
Effectors of the AvrE family are very large proteins of approximately 2000 amino acids, without sequence similarity with proteins of known function. Although their large size would be consistent with a multifunctional activity, their biochemical function remains unknown. However, the importance of this family of T3Es for pathogenicity on host plants and cell death elicitation on non‐host plants allowed us to test the functionality of mutated effector versions. Mutational analysis of Pa. stewartii WtsE showed that the full‐length protein is required for function (Ham et al., 2006, 2008). Consistent with this, we could not identify a subdomain of E. amylovora DspA/E sufficient to elicit cell death when transiently expressed (A. Degrave et al., unpublished observation). One exception is the first N‐terminal 200 amino acids dedicated to secretion and translocation, as described above, which could be removed from Pa. stewartii WtsE without affecting the cell death induction capacity of the transiently expressed construct (Ham et al., 2008). Following this N‐terminal secretion/translocation domain, protein‐threading software predicts a conserved double β‐propeller domain in all AvrE orthologues (Siamer et al., 2013). It is possible that this predicted domain acts as a platform‐binding domain for interactors (Chen and Chan, 2011).
Motif searches identified nuclear localization signals (NLSs), endoplasmic reticulum membrane retention signals (ERMRSs), leucine zipper motifs (LZs), peroxisome targeting signals (PTSs) and WXXXE motifs, but none of these motifs are present in all the AvrE effectors analysed. The ERMRS signal, when present, is always positioned in the penultimate four amino acids and its deletion impairs DspA/E or WtsE protein function (Ham et al., 2006, 2009; A. Degrave et al., unpublished observations). The position of other motifs is generally not conserved between orthologues (Ham et al., 2006; Siamer et al., 2013). WXXXE motifs are of particular interest, because they were also described in T3Es of animal‐pathogenic bacteria, which functionally mimic guanine nucleotide exchange factors (GEFs), the endogenous activators of Rho family GTPases (Orchard and Alto, 2012). Simultaneous mutagenesis of the two WXXXE motifs of Pa. stewartii WtsE alters the function of the protein (Ham et al., 2009). When the WXXXE motifs are not detected in orthologous proteins, as in Pe. carotovorum DspE or R. solanacearum PopS, tryptophan residues are nevertheless conserved at the same position (Hogan et al., 2013; Jacobs et al., 2013), and site‐directed mutagenesis of these two tryptophan residues is deleterious for the function of Pe. carotovorum DspE (Hogan et al., 2013). Therefore, it appears that at least these tryptophan residues are important to maintain the function of the protein. Whether these residues are important for correct folding of the protein or are involved in a putative catalytic GEF function remains to be determined.
Localization Inside the Eukaryotic Cell
Attempts to localize AvrE family effectors inside plant cells following Agrobacterium‐mediated transient expression were unsuccessful (Boureau et al., 2006; Degrave et al., 2013; Ham et al., 2009). This is probably because of the toxic mode of action of AvrE‐T3Es, which leads to a rapid repression of de novo protein synthesis (Degrave et al., 2013). A transient GFP signal has nevertheless been detected inside the plant nucleus in a transgenic A. thaliana line expressing a GFP–DspA/E fusion (Degrave et al., 2013). This is consistent with predictions made with the psortii program, which predict that all AvrE family effectors, except PopS from R. solanacearum, may localize to the plant cell nucleus (Ham et al., 2006). However, not all AvrE‐T3Es harbour an NLS signal, and mutagenesis of the Pa. stewartii WtsE and E. amylovora DspA/E NLS indicates that this motif is not required for protein function (Ham et al., 2006; A. Degrave et al., unpublished observations). Therefore, if the protein is transported inside the plant nucleus, this could be achieved without a functional NLS. It is also possible that the function of AvrE family effectors is not dependent on nucleus localization and may be linked to localization in another cellular compartment. AvrE‐like effectors may also localize to the endoplasmic reticulum because most effectors of the family harbour an ERMRS motif at their C‐terminus.
Yeast as a Tool to Unravel the Function of AvrE Family Effectors
Induction of the expression of E. amylovora DspA/E or Pa. stewartii WtsE in Saccharomyces cerevisiae leads to growth arrest (Ham et al., 2008; Meng et al., 2006; Oh et al., 2007; Siamer et al., 2011) and strongly affects cellular traffic (Siamer et al., 2011). This phenotype is reminiscent of the growth inhibition observed on A. thaliana transgenic lines expressing a low level of DspA/E (Degrave et al., 2013), and suggests that AvrE‐like effectors probably target a general pathway conserved in eukaryotes.
Yeast toxicity precludes yeast two‐hybrid experiments with full‐length effectors of the AvrE family. However, yeast two‐hybrid assays performed with protein fragments of Pa. stewartii WtsE and E. amylovora DspA/E identified leucine‐rich repeat receptor‐like serine/threonine kinases (RLKs) as putative interactors (Ham et al., 2006; Meng et al., 2006). Whether these interactions occur in vivo remains to be determined. Interactions of WtsE with two maize regulatory subunits of protein phosphatase 2A (PP2A) have also been reported in a recent review (Roper, 2011). Although this awaits publication of the data, it is tempting to speculate that these latter interactions occur in vivo, as it has been shown that E. amylovora DspA/E expression in yeast activates PP2A (Siamer et al., 2014). Activated PP2A dephosphorylates and activates Orm proteins, which are the negative regulators of serine palmitoyltransferase (SPT), the first enzyme of the sphingolipid pathway (Breslow et al., 2010; Sun et al., 2012). This, in turn, leads to the inhibition of SPT activity and decreases the level of long‐chain bases (LCBs), the products of SPT activity. Feeding yeast cells expressing DspA/E with LCB, or expressing DspA/E in yeast mutants that accumulate LCB, rescues yeast cell growth and cellular traffic, indicating that DspA/E toxicity is linked to LCB depletion (Siamer et al., 2014). Inhibition of SPT could explain all the toxic phenotypes observed. Indeed, SPT inhibition leads to the perturbation of cellular traffic (Zanolari et al., 2000), inhibition of translation initiation (Meier et al., 2006), growth arrest and, ultimately, cell death (Dickson et al., 2006; Pinto et al., 1992).
Given the functional complementation observed between effectors of the AvrE family (Bogdanove et al., 1998a; Ham et al., 2006; Kvitko et al., 2009), it is likely that the expression of other members of the AvrE family could also activate PP2A and decrease the LCB level, but this remains to be tested.
Future Prospects
Yeast studies allow the proposal of several non‐exclusive hypotheses to explain how AvrE‐T3Es could inhibit PTI (Fig 1). First, yeast two‐hybrid studies indicate that effectors of the AvrE family could interact with RLKs (Ham et al., 2006; Meng et al., 2006). As the predicted structure of these RLKs is similar to a class of surface‐localized immune receptors (Afzal et al., 2008; Dangl and Jones, 2001), a possible consequence of direct interactions with these RLKs of host plants during infection could be to block signal transduction leading to defence responses. Effectors of the AvrE family might also interact with and activate PP2A (Roper, 2011; Siamer et al., 2014). Activation of PP2A by effectors of the AvrE family would reinforce PTI inhibition as PP2A is involved in the negative control of PTI in planta (Segonzac et al., 2014; Trotta et al., 2011). The involvement of several putative substrates of PP2A could now be tested in planta following bacterial infection. Yeast studies indicate that ORM proteins, which negatively regulate SPT activity and sphingolipid biosynthesis, could be important PP2A substrates in planta (Siamer et al., 2014). This would make sense as the sphingolipid biosynthetic pathway is induced during the hypersensitive response that blocks pathogen attack at the site of infection (Berkey et al., 2012; Peer et al., 2010; Shi et al., 2007). Therefore, inhibition of this pathway could be seen as a mechanism that delays hypersensitive response cell death and allows bacterial development in planta. Perturbation of the sphingolipid pathway in planta could also explain why effectors of the AvrE family induce a slow plant cell death, because sphingolipids are structural lipids of plant membranes and prolonged inhibition of the sphingolipid pathway ultimately leads to plant cell death (Chen et al., 2006). Another substrate involved in PTI that could be targeted following AvrE‐mediated activation of PP2A in planta is BAK1, whose steady‐state phosphorylation is important to trigger a defence reaction and whose dephosphorylation requires PP2A (Segonzac et al., 2014). Overall, yeast studies suggest that effectors of the AvrE family could reinforce PTI inhibition at different levels. This remains to be tested in planta with different members of the family. In particular, as most of the studies were performed with AvrE (pseudomonads) and DspA/E, DspE or WtsE (enterobacteria), it would be very interesting to check whether PopS (Ralstonia spp.), HopR1 (pseudomonads) and XopAM (xanthomonads), which are more distantly related, function similarly.
Figure 1.
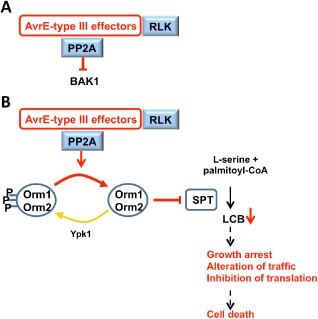
Proposed model for AvrE‐like effector‐mediated toxicity and inhibition of pathogen‐associated molecular pattern (PAMP)‐triggered immunity (PTI deduced from yeast studies. Yeast two‐hybrid studies indicate that AvrE‐type III effectors (T3Es) possibly interact with receptor‐like serine/threonine kinases (RLKs) and protein phosphatase 2A (PP2A) (Ham et al., 2006; Meng et al., 2006; Roper, 2011). Direct binding to RLKs could potentially block signal transduction, leading to plant defence (Meng et al., 2006). (A) Binding and activation of PP2A could also reinforce PTI inhibition indirectly through dephosphorylation and inactivation of BAKI (Segonzac et al., 2014; Siamer et al., 2014). As RLKs and BAK1 are localized to the plasma membrane (PM), PM is a potential site of action for AvrE‐T3Es. (B) Once activated, PP2A could also dephosphorylate and activate Orm proteins. As ORM proteins are associated with serine palmitoyltransferase (SPT) at the endoplasmic reticulum (ER), the ER is another potential site of action of AvrE‐T3Es. Dephosphorylated Orm proteins could, in turn, inhibit SPT, leading to long‐chain base (LCB) depletion. As the sphingolipid pathway is induced during the hypersensitive reaction (HR), inhibition of this pathway could delay HR cell death and participate in PTI inhibition (Berkey et al., 2012). Prolonged inhibition of the sphingolipid pathway will ultimately lead to plant cell death (Chen et al., 2006), explaining the ‘slow death’ phenotype observed on host and non‐host plants.
Beyond the role of the AvrE effector family during plant bacterial pathogenesis, it is interesting to note that several bacteria which associate with plants, but which are not reported to be pathogens, also harbour an effector of the AvrE family. For example, Marinomonas mediterannea, a bacterium reported to associate with sea grass, harbours a gene showing 26% identity with DspA/E of E. amylovora (Siamer et al., 2013). The same is true of P. fluorescens SBW25, which harbours DspE, although it is non‐pathogenic and does not elicit an HR in any host plant tested (Preston et al., 2001). As the low expression of effectors of the AvrE family is not associated with plant cell death (Degrave et al., 2013), it would be interesting to determine whether a low expression level of effectors of the AvrE family could be beneficial for the association of these non‐pathogenic bacteria with plants.
Finally, an intriguing question is the origin of the AvrE family of T3Es. Until now, no homology to any known protein has been reported. Interestingly, T3Es of the AvrE family and several viruses inhibit the whole sphingolipid pathway of their hosts (Rosenwasser et al., 2014). This could be a case of convergent evolution; however, it would be interesting to determine whether T3Es of the AvrE family share similarity with a viral protein.
Acknowledgements
We thank Jacques Pédron for critical reading of the manuscript. This review is dedicated to the memory of Roland Chartier.
References
- Afzal, A.J. , Wood, A.J. and Lightfoot, D.A. (2008) Plant receptor‐like serine threonine kinases: roles in signaling and plant defense. Mol. Plant–Microbe Interact. 21, 507–517. [DOI] [PubMed] [Google Scholar]
- Badel, J.L. , Shimizu, R. , Oh, H.‐S. and Collmer, A. (2006) A Pseudomonas syringae pv. tomato avrE1/hopM1 mutant is severely reduced in growth and lesion formation in tomato. Mol. Plant–Microbe Interact. 19, 99–111. [DOI] [PubMed] [Google Scholar]
- Barash, I. and Manulis‐Sasson, S. (2007) Virulence mechanisms and host specificity of gall‐forming Pantoea agglomerans . Trends Microbiol. 15, 538–545. [DOI] [PubMed] [Google Scholar]
- Berkey, R. , Bendigeri, D. and Xiao, S. (2012) Sphingolipids and plant defense/disease: the ‘death’ connection and beyond. Front. Plant Sci. 3, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block, A. and Alfano, J.R. (2011) Plant targets for Pseudomonas syringae type III effectors: virulence targets or guarded decoys? Curr. Opin. Microbiol. 14, 39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocsanczy, A.M. , Nissinen, R.M. , Oh, C.‐S. and Beer, S.V. (2008) HrpN of Erwinia amylovora functions in the translocation of DspA/E into plant cells. Mol. Plant Pathol. 9, 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanove, A.J. , Kim, J.F. , Wei, Z. , Kolchinsky, P. , Charkowski, A.O. , Conlin, A.K. , Collmer, A. and Beer, S.V. (1998a) Homology and functional similarity of an hrp‐linked pathogenicity locus, dspEF, of Erwinia amylovora and the avirulence locus avrE of Pseudomonas syringae pathovar tomato . Proc. Natl. Acad. Sci. USA, 95, 1325–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanove, A.J. , Bauer, D.W. and Beer, S.V. (1998b) Erwinia amylovora secretes DspE, a pathogenicity factor and functional AvrE homolog, through the Hrp (type III secretion) pathway. J. Bacteriol. 180, 2244–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boureau, T. , ElMaarouf‐Bouteau, H. , Garnier, A. , Brisset, M.‐N. , Perino, C. , Pucheu, I. and Barny, M.‐A. (2006) DspA/E, a type III effector essential for Erwinia amylovora pathogenicity and growth in planta, induces cell death in host apple and nonhost tobacco plants. Mol. Plant–Microbe Interact. 19, 16–24. [DOI] [PubMed] [Google Scholar]
- Boureau, T. , Siamer, S. , Perino, C. , Gaubert, S. , Patrit, O. , Degrave, A. , Fagard, M. , Chevreau, E. and Barny, M.‐A. (2011) The HrpN effector of Erwinia amylovora, which is involved in type III translocation, contributes directly or indirectly to callose elicitation on apple leaves. Mol. Plant–Microbe Interact. 24, 577–584. [DOI] [PubMed] [Google Scholar]
- Breslow, D.K. , Collins, S.R. , Bodenmiller, B. , Aebersold, R. , Simons, K. , Shevchenko, A. , Ejsing, C.S. and Weissman, J.S. (2010) Orm family proteins mediate sphingolipid homeostasis. Nature, 463, 1048–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.K.M. and Chan, N.L. (2011) The many blades of the β‐propeller proteins: conserved but versatile. Trends Biochem. Sci. 36, 553–561. [DOI] [PubMed] [Google Scholar]
- Chen, M. , Han, G. , Dietrich, C.R. , Dunn, T.M. and Cahoon, E.B. (2006) The essential nature of sphingolipids in plants as revealed by the functional identification and characterization of the Arabidopsis LCB1 subunit of serine palmitoyltransferase. Plant Cell, 18, 3576–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm, S.T. , Coaker, G. , Day, B. and Staskawicz, B.J. (2006) Host–microbe interactions: shaping the evolution of the plant immune response. Cell, 124, 803–814. [DOI] [PubMed] [Google Scholar]
- Cunnac, S. , Chakravarthy, S. , Kvitko, B.H. , Russell, A.B. , Martin, G.B. and Collmer, A. (2011) Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae . Proc. Natl. Acad. Sci. USA, 108, 2975–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J.L. and Jones, J.D. (2001) Plant pathogens and integrated defence responses to infection. Nature, 411, 826–833. [DOI] [PubMed] [Google Scholar]
- DebRoy, S. , Thilmony, R. , Kwack, Y.‐B. , Nomura, K. and He, S.Y. (2004) A family of conserved bacterial effectors inhibits salicylic acid‐mediated basal immunity and promotes disease necrosis in plants. Proc. Natl. Acad. Sci. USA, 101, 9927–9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degrave, A. , Moreau, M. , Launay, A. , Barny, M.‐A. , Brisset, M.‐N. , Patrit, O. , Taconnat, L. , Vedel, R. and Fagard, M. (2013) The bacterial effector DspA/E is toxic in Arabidopsis thaliana and is required for multiplication and survival of fire blight pathogen: DspA/E is required for Ea growth in A. thaliana . Mol. Plant Pathol. 14, 506–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson, R.C. , Sumanasekera, C. and Lester, R.L. (2006) Functions and metabolism of sphingolipids in Saccharomyces cerevisiae . Prog. Lipid Res. 45, 447–465. [DOI] [PubMed] [Google Scholar]
- Frederick, R.D. , Ahmad, M. , Majerczak, D.R. , Arroyo‐Rodríguez, A.S. , Manulis, S. and Coplin, D.L. (2001) Genetic organization of the Pantoea stewartii subsp. stewartii hrp gene cluster and sequence analysis of the hrpA, hrpC, hrpN, and wtsE operons. Mol. Plant–Microbe Interact. 14, 1213–1222. [DOI] [PubMed] [Google Scholar]
- Gaudriault, S. , Malandrin, L. , Paulin, J. and P., and Barny, M.‐A. (1997) DspA, an essential pathogenicity factor of Erwinia amylovora showing homology with AvrE of Pseudomonas syringae, is secreted via the Hrp secretion pathway in a DspB‐dependent way. Mol. Microbiol. 26, 1057–1069. [DOI] [PubMed] [Google Scholar]
- Gaudriault, S. , Paulin, J.‐P. and Barny, M.‐A. (2002) The DspB/F protein of Erwinia amylovora is a type III secretion chaperone ensuring efficient intrabacterial production of the Hrp‐secreted DspA/E pathogenicity factor. Mol. Plant Pathol. 3, 313–320. [DOI] [PubMed] [Google Scholar]
- Glasner, J.D. , Yang, C.‐H. , Reverchon, S. , Hugouvieux‐Cotte‐Pattat, N. , Condemine, G. , Bohin, J. , Van Gijsegem, F. , Yang, S. , Franza, T. , Expert, D. , Plunkett, G. , San Francisco, M.J. , Charkowski, A.O. , Py, B. , Bell, , , K. , Rauscher, L. , Rodriguez‐Palenzuela, P. , Toussaint, A. , Holeva, M.C. , He, S.Y. , Douet, V. , Boccara, M. , Blanco, C. , Toth, I. , Anderson, B.D. , Biehl, B.S. , Mau, B. , Flynn, S.M. , Barras, F. , Lindeberg, M. , Birch, P.R. , Tsuyumu, S. , Shi, X. , Hibbing, M. , Yap, M.N. , Carpentier, M. , Dassa, E. , Umehara, M. , Kim, J.F. , Rusch, , , M. , Soni, P. , Mayhew, G.F. , Fouts, D.E. , Gill, S.R. , Blattner, F.R. , Keen, N.T. and Perna, N.T. (2011) Genome sequence of the plant‐pathogenic bacterium Dickeya dadantii 3937. J. Bacteriol. 193, 2076–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham, J.H. , Majerczak, D.R. , Arroyo‐Rodriguez, A.S. , Mackey, D.M. and Coplin, D.L. (2006) WtsE, an AvrE‐family effector protein from Pantoea stewartii subsp. stewartii, causes disease‐associated cell death in corn and requires a chaperone protein for stability. Mol. Plant–Microbe Interact. 19, 1092–1102. [DOI] [PubMed] [Google Scholar]
- Ham, J.H. , Majerczak, D. , Ewert, S. , Sreerekha, M.‐V. , Mackey, D. and Coplin, D. (2008) WtsE, an AvrE‐family type III effector protein of Pantoea stewartii subsp. stewartii, causes cell death in non‐host plants. Mol. Plant Pathol. 9, 633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham, J.H. , Majerczak, D.R. , Nomura, K. , Mecey, C. , Uribe, F. , He, S.‐Y. , Mackey, D. and Coplin, D.L. (2009) Multiple activities of the plant pathogen type III effector proteins WtsE and AvrE require WxxxE motifs. Mol. Plant–Microbe Interact. 22, 703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan, C.S. , Mole, B.M. , Grant, S.R. , Willis, D.K. and Charkowski, A.O. (2013) The type III secreted effector DspE is required early in Solanum tuberosum leaf infection by Pectobacterium carotovorum to cause cell death, and requires Wx(3–6)D/E motifs. PLoS ONE, 8, e65534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holeva, M.C. , Bell, K.S. , Hyman, L.J. , Avrova, A.O. , Whisson, S.C. , Birch, P.R.J. and Toth, I.K. (2004) Use of a pooled transposon mutation grid to demonstrate roles in disease development for Erwinia carotovora subsp. atroseptica putative type III secreted effector (DspE/A) and helper (HrpN) proteins. Mol. Plant–Microbe Interact. 17, 943–950. [DOI] [PubMed] [Google Scholar]
- Jacobs, J.M. , Milling, A. , Mitra, R.M. , Hogan, C.S. , Ailloud, F. , Prior, P. and Allen, C. (2013) Ralstonia solanacearum requires PopS, an ancient AvrE‐family effector, for virulence and to overcome salicylic acid‐mediated defenses during tomato pathogenesis. Mbio, 4, 1–12. e00875–e00813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W. , Jiang, B.‐L. , Xu, R.‐Q. , Huang, J.‐D. , Wei, H.‐Y. , Jiang, G.‐F. , Cen, W.‐J. , Liu, J. , Ge, Y.‐Y. , Li, G.‐H. , Su, L.L. , Hang, X.H. , Tang, D.J. , Lu, G.T. , Feng, J.X. , He, Y.Q. and Tang, J.L. (2009) Identification of six type III effector genes with the PIP box in Xanthomonas campestris pv. campestris and five of them contribute individually to full pathogenicity. Mol. Plant–Microbe Interact. 22, 1401–1411. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kim, H.‐S. , Thammarat, P. , Lommel, S.A. , Hogan, C.S. and Charkowski, A.O. (2011) Pectobacterium carotovorum elicits plant cell death with DspE/F but the P. carotovorum DspE does not suppress callose or induce expression of plant genes early in plant–microbe interactions. Mol. Plant–Microbe Interact. 24, 773–786. [DOI] [PubMed] [Google Scholar]
- Kobayashi, D.Y. , Tamaki, S.J. and Keen, N.T. (1989) Cloned avirulence genes from the tomato pathogen Pseudomonas syringae pv. tomato confer cultivar specificity on soybean. Proc. Natl. Acad. Sci. USA, 86, 157–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvitko, B.H. , Park, D.H. , Velásquez, A.C. , Wei, C.‐F. , Russell, A.B. , Martin, G.B. , Schneider, D.J. and Collmer, A. (2009) Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 5, e1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeberg, M. , Cunnac, S. and Collmer, A. (2012) Pseudomonas syringae type III effector repertoires: last words in endless arguments. Trends Microbiol. 20, 199–208. [DOI] [PubMed] [Google Scholar]
- Lorang, J.M. and Keen, N.T. (1995) Characterization of avrE from Pseudomonas syringae pv. tomato: a hrp‐linked avirulence locus consisting of at least two transcriptional units. Mol. Plant–Microbe Interact. 8, 49–57. [DOI] [PubMed] [Google Scholar]
- Luna, E. , Pastor, V. , Robert, J. , Flors, V. , Mauch‐Mani, B. and Ton, J. (2011) Callose deposition: a multifaceted plant defense response. Mol. Plant–Microbe Interact. 24, 183–193. [DOI] [PubMed] [Google Scholar]
- McNally, R.R. , Toth, I.K. , Cock, P.J.A. , Pritchard, L. , Hedley, P.E. , Morris, J.A. , Zhao, Y. and Sundin, G.W. (2012) Genetic characterization of the HrpL regulon of the fire blight pathogen Erwinia amylovora reveals novel virulence factors: the HrpL regulon of Erwinia amylovora . Mol. Plant Pathol. 13, 160–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier, K.D. , Deloche, O. , Kajiwara, K. , Funato, K. and Riezman, H. (2006) Sphingoid base is required for translation initiation during heat stress in Saccharomyces cerevisiae . Mol. Biol. Cell, 17, 1164–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, X. , Bonasera, J.M. , Kim, J.F. , Nissinen, R.M. and Beer, S.V. (2006) Apple proteins that interact with DspA/E, a pathogenicity effector of Erwinia amylovora, the fire blight pathogen. Mol. Plant–Microbe Interact. 19, 53–61. [DOI] [PubMed] [Google Scholar]
- Mor, H. , Manulis, S. , Zuck, M. , Nizan, R. , Coplin, D.L. and Barash, I. (2001) Genetic organization of the hrp gene cluster and dspAE/BF operon in Erwinia herbicola pv. gypsophilae . Mol. Plant–Microbe Interact. 14, 431–436. [DOI] [PubMed] [Google Scholar]
- Oh, C.S. , Carpenter, S.C.D. , Hayes, M.L. and Beer, S.V. (2010) Secretion and translocation signals and DspB/F‐binding domains in the type III effector DspA/E of Erwinia amylovora . Microbiology, 156, 1211–1220. [DOI] [PubMed] [Google Scholar]
- Oh, C.‐S. , Martin, G.B. and Beer, S.V. (2007) DspA/E, a type III effector of Erwinia amylovora, is required for early rapid growth in Nicotiana benthamiana and causes NbSGT1‐dependent cell death. Mol. Plant Pathol. 8, 255–265. [DOI] [PubMed] [Google Scholar]
- Orchard, R.C. and Alto, N.M. (2012) Mimicking GEFs: a common theme for bacterial pathogens: bacterial GEF mimics. Cell, 14, 10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer, M. , Stegmann, M. , Mueller, M.J. and Waller, F. (2010) Pseudomonas syringae infection triggers de novo synthesis of phytosphingosine from sphinganine in Arabidopsis thaliana . FEBS Lett. 584, 4053–4056. [DOI] [PubMed] [Google Scholar]
- Peeters, N. , Carrère, S. , Anisimova, M. , Plener, L. , Cazalé, A.C. and Genin, S. (2013) Repertoire, unified nomenclature and evolution of the type III effector gene set in the Ralstonia solanacearum species complex. BMC Genomics, 6, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Q. , Yang, S. , Charkowski, A.O. , Yap, M.‐N. , Steeber, D.A. , Keen, N.T. and Yang, C.‐H. (2006) Population behavior analysis of dspE and pelD regulation in Erwinia chrysanthemi 3937. Mol. Plant–Microbe Interact. 19, 451–457. [DOI] [PubMed] [Google Scholar]
- Pester, D. , Milčevičová, R. , Schaffer, J. , Wilhelm, E. and Blümel, S. (2012) Erwinia amylovora expresses fast and simultaneously hrp/dsp virulence genes during flower infection on apple trees. PLoS ONE, 7, e32583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto, W.J. , Srinivasan, B. , Shepherd, S. , Schmidt, A. , Dickson, R.C. and Lester, R.L. (1992) Sphingolipid long‐chain‐base auxotrophs of Saccharomyces cerevisiae: genetics, physiology, and a method for their selection. J. Bacteriol. 174, 2565–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston, G.M. , Bertrand, N. and Rainey, P.B. (2001) Type III secretion in plant growth‐promoting Pseudomonas fluorescens SBW25. Mol. Microbiol. 41, 999–1014. [DOI] [PubMed] [Google Scholar]
- Qian, W. , Jia, Y. , Ren, S.‐X. , He, Y.‐Q. , Feng, J.‐X. , Lu, L.‐F. , Sun, Q. , Ying, G. , Tang, D.‐J. , Tang, H. , Wu, W. , Hao, P. , Wang, L. , Jiang, B.L. , Zeng, S. , Gu, W.Y. , Lu, G. , Rong, L. , Tian, Y. , Yao, Z. , Fu, G. , Chen, B. , Fang, R. , Qiang, B. , Chen, Z. , Zhao, G.P. , Tang, J.L. and He, C. (2005) Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv. campestris . Genome Res. 15, 757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer, L. , Guttman, D.S. and Dangl, J.L. (2004) Diverse evolutionary mechanisms shape the type III effector virulence factor repertoire in the plant pathogen Pseudomonas syringae . Genetics, 167, 1341–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper, M.C. (2011) Pantoea stewartii subsp. stewartii: lessons learned from a xylem‐dwelling pathogen of sweet corn: Pantoea stewartii subsp. stewartii. Mol. Plant Pathol. 12, 628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwasser, S. , Mausz, M.A. , Schatz, D. , Sheyn, U. , Malitsky, S. , Aharoni, A. , Weinstock, E. , Tzfadia, O. , Ben‐Dor, S. , Feldmesser, E. , Pohnert, G. and Vardi, A. (2014) Rewiring host lipid metabolism by large viruses determines the fate of Emiliania huxleyi, a bloom‐forming alga in the ocean. Plant Cell, 26, 2689–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonzac, C. , Macho, A.P. , Sanmartín, M. , Ntoukakis, V. , Sánchez‐Serrano, J.J. and Zipfel, C. (2014) Negative control of BAK1 by protein phosphatase 2A during plant innate immunity. EMBO J. 33, 2069–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, L. , Bielawski, J. , Mu, J. , Dong, H. , Teng, C. , Zhang, J. , Yang, X. , Tomishige, N. , Hanada, K. , Hannun, Y.A. and Zuo, J. (2007) Involvement of sphingoid bases in mediating reactive oxygen intermediate production and programmed cell death in Arabidopsis. Cell Res. 17, 1030–1040. [DOI] [PubMed] [Google Scholar]
- Siamer, S. , Patrit, O. , Fagard, M. , Belgareh‐Touzé, N. and Barny, M.A. (2011) Expressing the Erwinia amylovora type III effector DspA/E in the yeast Saccharomyces cerevisiae strongly alters cellular trafficking. FEBS Open Biol. 1, 23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siamer, S. , Gaubert, S. , Boureau, T. , Brisset, M.‐N. and Barny, M.‐A. (2013) Mutational analysis of a predicted double β‐propeller domain of the DspA/E effector of Erwinia amylovora . FEMS Microbiol. Lett. 342, 54–61. [DOI] [PubMed] [Google Scholar]
- Siamer, S. , Guillas, I. , Shimobayashi, M. , Kunz, C. , Hall, M.N. and Barny, M.‐A. (2014) Expression of the bacterial type III effector DspA/E in Saccharomyces cerevisiae down‐regulates the sphingolipid biosynthetic pathway leading to growth arrest. J. Biol. Chem. 289, 18 466–18 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y. , Miao, Y. , Yamane, Y. , Zhang, C. , Shokat, K.M. , Takematsu, H. , Kozutsumi, Y. and Drubin, D.G. (2012) Orm protein phosphoregulation mediates transient sphingolipid biosynthesis response to heat stress via the Pkh‐Ypk and Cdc55‐PP2A pathways. Mol. Biol. Cell, 23, 2388–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triplett, L.R. , Melotto, M. and Sundin, G.W. (2009) Functional analysis of the N terminus of the Erwinia amylovora secreted effector DspA/E reveals features required for secretion, translocation, and binding to the chaperone DspB/F. Mol. Plant–Microbe Interact. 22, 1282–1292. [DOI] [PubMed] [Google Scholar]
- Triplett, L.R. , Wedemeyer, W.J. and Sundin, G.W. (2010) Homology‐based modeling of the Erwinia amylovora type III secretion chaperone DspF used to identify amino acids required for virulence and interaction with the effector DspE. Res. Microbiol. 161, 613–618. [DOI] [PubMed] [Google Scholar]
- Trotta, A. , Wrzaczek, M. , Scharte, J. , Tikkanen, M. , Konert, G. , Rahikainen, M. , Holmström, M. , Hiltunen, H.M. , Rips, S. , Sipari, N. , Mulo, P. , Weis, E. , von Schaewen, A. , Aro, E.M. and Kangasjärvi, S. (2011) Regulatory subunit B′gamma of protein phosphatase 2A prevents unnecessary defense reactions under low light in Arabidopsis. Plant Physiol. 156, 1464–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D. , Korban, S.S. and Zhao, Y. (2010) Molecular signature of differential virulence in natural isolates of Erwinia amylovora . Phytopathology, 100, 192–198. [DOI] [PubMed] [Google Scholar]
- Wei, Z.M. , Sneath, B.J. and Beer, S.V. (1992) Expression of Erwinia amylovora hrp genes in response to environmental stimuli. J. Bacteriol. 174, 1875–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S. , Perna, N.T. , Cooksey, D.A. , Okinaka, Y. , Lindow, S.E. , Ibekwe, A.M. , Keen, N.T. and Yang, C.‐H. (2004) Genome‐wide identification of plant‐upregulated genes of Erwinia chrysanthemi 3937 using a GFP‐based IVET leaf array. Mol. Plant–Microbe Interact. 17, 999–1008. [DOI] [PubMed] [Google Scholar]
- Yang, S. , Peng, Q. , Zhang, Q. , Zou, L. , Li, Y. , Robert, C. , Pritchard, L. , Liu, H. , Hovey, R. , Wang, Q. , Birch, P. , Toth, I.K. and Yang, C.H. (2010) Genome‐wide identification of HrpL‐regulated genes in the necrotrophic phytopathogen Dickeya dadantii 3937. PLoS ONE, 5, e13472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanolari, B. , Friant, S. , Funato, K. , Sütterlin, C. , Stevenson, B.J. and Riezman, H. (2000) Sphingoid base synthesis requirement for endocytosis in Saccharomyces cerevisiae . EMBO J. 19, 2824–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]


