Figure 1.
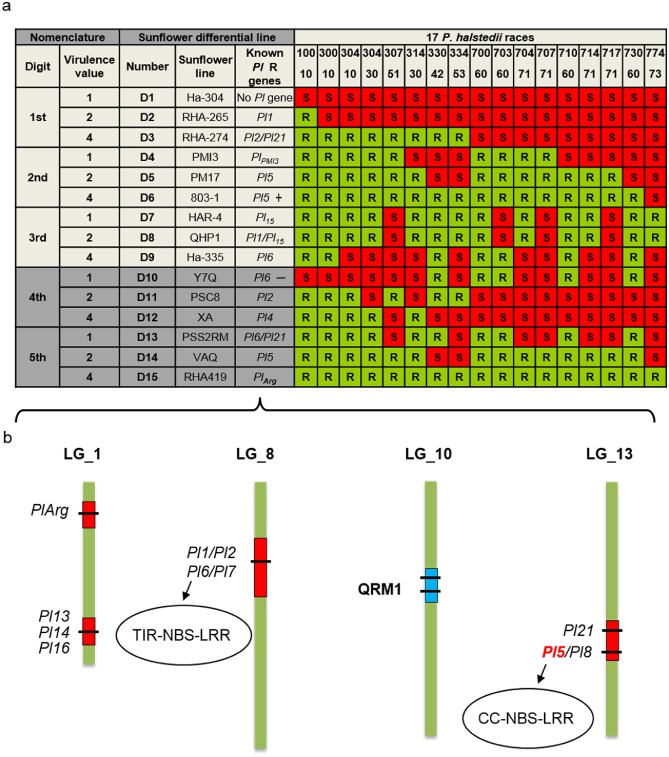
Nomenclature of Plasmopara halstedii pathotypes and genetic map of sunflower resistance genes. (a) Plasmopara halstedii pathotypes were defined according to an international nomenclature based on the virulence profile of a given isolate on 15 sunflower differential lines (D1–D15) selected according to their resistance patterns (Gulya et al., 1998; Tourvieille de Labrouhe et al., 2012). Susceptibility (S) and resistance (R) were defined by the presence or absence of disease symptoms and sporulation on leaves 2–3 weeks after inoculation of sunflower root seedlings, grown in soil (Mouzeyar et al., 1993). A triplet coding system was initially set up on nine sunflower lines (D1–D9, Gulya et al., 1998), but the occurrence of new P. halstedii pathotypes led to the addition of six other sunflower lines (in dark grey) and to a five‐digit coding system (D10–D15, Tourvieille de Labrouhe et al., 2012). The phenotyping results on each triplet of sunflower differential lines give the pathotype digit values. If the first differential line of a set of three is susceptible, a value of ‘1’ is assigned to the pathotype. If the second line is S, a value of ‘2’, and, for the third line, a value of ‘4’. When the line is resistant, a value of ‘0’ is assigned to the pathotype. The virulence code is additive within each set. For example, virulence code 710 is explained by ‘7’ (S for D1–D3, 1 + 2 + 4 = 7), ‘1’ (S for D4) and ‘0’ (R for D7–D9). Dominant genes giving resistance to P. halstedii are called Pl genes. The known functional R genes are indicated in the sunflower differential lines. ‘+’ or ‘–’ after the Pl gene indicates the strength of resistance alleles. (b) Eleven sunflower Pl genes have been mapped on three of 17 sunflower linkage groups (LGs) (Bachlava et al., 2011; Bouzidi et al., 2002; Franchel et al., 2013; Liu et al., 2012; Mulpuri et al., 2009; Radwan et al., 2003; Vincourt et al., 2012; Wieckhorst et al., 2010). Pl1/Pl2/Pl6 and Pl7 are localized in a Toll/interleukin‐1 receptor‐nucleotide‐binding site‐leucine‐rich repeat (TIR‐NBS‐LRR)‐rich region. Pl5/Pl8 and Pl21 are localized in a coiled‐coil (CC)‐NBS‐LRR‐rich region. QRM1, the major quantitative trait locus (QTL), conferring a quantitative resistance to pathotype 710 of P. halstedii, was mapped on LG10 (Vear et al., 2008b; Vincourt et al., 2012).
