Figure 1.
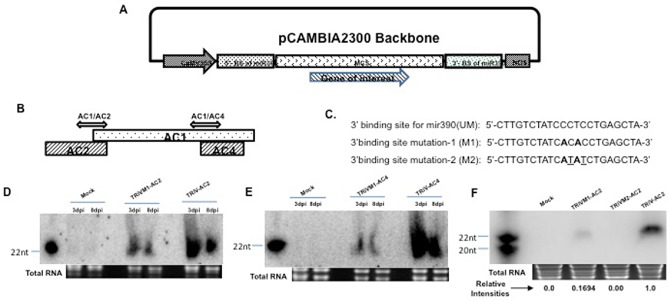
Cloning and expression of TRiV‐AC2 and TRiV‐AC4 in tobacco. (A) Map showing the TRiV construct containing 5′‐ and 3′‐binding sites of miR390 and the multiple cloning site (MCS) in which the sequence of interest can be cloned. (B) Schematic representation of DNA‐A showing the positions of AC1, AC2 and AC4 open reading frames (ORFs). Double‐headed arrows represent the fragments used for cloning in TRiV. (C) Sequences of wild‐type (UM) and mutated 3′‐binding sites of miR390. The mutated nucleotides are denoted in bold letters in M1 and additional mutated nucleotides are underlined in M2. (D) Northern blot analyses performed at 3 and 8 days post‐infiltration (dpi) of small interfering RNA (siRNA) from tobacco leaves infiltrated with Mock (empty vector), mutated (TRiVM1‐AC2) and TRiV‐AC2. (E) Same analyses with Mock, TRiVM1‐AC4 and TRiV‐AC4, as indicated above the lanes. Polymerase chain reaction (PCR)‐amplified AC1/AC2 and AC1/AC4 fragments (C) were taken as probes for TRiV‐AC2 and TRiV‐AC4, respectively. AC2SM22 and AC4SM22 were used as size markers for (D) and (E), respectively. Loading controls are indicated by total RNA below each panel. (F) Northern blot of siRNA at 8 dpi in plants infiltrated with TRiV‐AC2 with its mutants (TRiVM1 and TRiVM2). The relative intensities of the bands are presented below. AC2SM20 and AC2SM22 were used as size markers for 20 and 22 nucleotides (nt), respectively.
