SUMMARY
The genomes of many filamentous fungi consist of a ‘core’ part containing conserved genes essential for normal development as well as conditionally dispensable (CD) or lineage‐specific (LS) chromosomes. In the plant‐pathogenic fungus Fusarium oxysporum f. sp. lycopersici, one LS chromosome harbours effector genes that contribute to pathogenicity. We employed flow cytometry to select for events of spontaneous (partial) loss of either the two smallest LS chromosomes or two different core chromosomes. We determined the rate of spontaneous loss of the ‘effector’ LS chromosome in vitro at around 1 in 35 000 spores. In addition, a viable strain was obtained lacking chromosome 12, which is considered to be a part of the core genome. We also isolated strains carrying approximately 1‐Mb deletions in the LS chromosomes and in the dispensable core chromosome. The large core chromosome 1 was never observed to sustain deletions over 200 kb. Whole‐genome sequencing revealed that some of the sites at which the deletions occurred were the same in several independent strains obtained for the two chromosomes tested, indicating the existence of deletion hotspots. For the core chromosome, this deletion hotspot was the site of insertion of the marker used to select for loss events. Loss of the core chromosome did not affect pathogenicity, whereas loss of the effector chromosome led to a complete loss of pathogenicity.
Keywords: chromosome deletions, conditionally dispensable chromosomes, flow cytometry, pathogenic fungi
Introduction
Lineage‐specific (LS) chromosomes are not present in all strains of a species and share no synteny with closely related species (Covert, 1998). In some cases, LS chromosomes have been shown to be lost without affecting growth, and are then called conditionally dispensable (CD) chromosomes. As these terms (LS and CD) are often used interchangeably in the literature, we follow the term commonly used for each fungus and then clarify our own use of these terms.
Comparative genomics and other approaches have led to the discovery and characterization of LS chromosomes in a number of plant‐pathogenic filamentous fungi, including Fusarium oxysporum (Ma et al., 2010), Nectria haematococca (Coleman et al., 2009), Leptosphaeria maculans (Rouxel et al., 2011), Alternaria alternata and Alternaria arborescens (Hatta et al., 2002; Hu et al., 2012), and Mycosphaerella graminicola (Goodwin et al., 2011). In several cases, there is a direct link between pathogenicity and LS chromosomes. In F. oxysporum f. sp. lycopersici strain Fol4287, genes encoding small proteins secreted in xylem (Six proteins or effectors) are located on an LS chromosome that can be horizontally transferred from one strain to another (Ma et al., 2010). LS chromosomes in this species are enriched for transposable elements and repeats.
The first discovery of CD chromosomes in N. haematococca dates back almost 25 years (Miao et al., 1991). Karyotyping showed these chromosomes to be meiotically unstable. Meiotic instability of CD chromosomes has also been described in L. maculans (Balesdent et al., 2013) and M. graminicola (Wittenberg et al., 2009). In L. maculans, CD chromosomes are lost in approximately 5% of the progeny from sexual crosses. In M. graminicola, this percentage is as high as 15%–20%. Loss of CD chromosomes during vegetative growth in N. haematococca can be induced by genetic transformation or treatment with the fungicide benomyl (VanEtten et al., 1998; Wasmann and VanEtten, 1996). Spontaneous loss during vegetative growth has been observed in A. alternata (Johnson et al., 2001) and in the insect‐infecting Metarhizium anisopliae (Wang et al., 2003).
LS or CD chromosomes have been suggested to constitute an accessory genome, with an elevated level of variation in the accumulation of non‐synonymous mutations and by loss or duplication of regions. This division in a core and accessory genome has been suggested to allow the fungal population to adapt more quickly to changing demands placed on it by the host (Croll and McDonald, 2012; Raffaele and Kamoun, 2012).
In this study, we investigated the stability of several LS and core (conserved) chromosomes in a tomato‐infecting strain of F. oxysporum, one of the top 10 fungal pathogens in molecular plant pathology, as voted by researchers in the field based on economic and scientific importance (Dean et al., 2012). We used the tomato‐infecting strain Fol4287, which contains a number of small and large LS chromosomes (Fig. 1). LS chromosome 14 carries almost all the genes for small secreted proteins found in the xylem sap of infected tomato plants (Ma et al., 2010; Schmidt et al., 2013).
Figure 1.
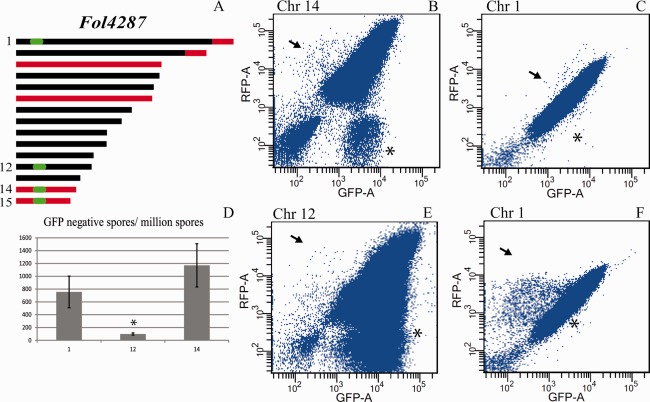
Selection for loss of expression of a marker gene from selected chromosomes. (A) Schematic representation of core (black) and lineage‐specific (LS, red) parts of the genome of Fol4287. Locations targeted for GFP (green fluorescent protein gene) insertion are indicated in green. (B, C, E, F) Dot plots showing flow cytometry analysis of strains containing both GFP and RFP (red fluorescent protein gene). Axis labels indicate the detection channel used. Axis scale values are given in arbitrary units that were defined to fit the range of fluorescence in the biological samples. Each panel displays 1 × 106 spores of a single culture with the GFP insertion in core chromosome 1 or 12 or LS chromosome 14 of Fol4287. Arrows indicate populations of spores that have lost GFP but not RFP, indicative of the loss of the associated genomic region. Asterisks indicate the populations that have lost RFP but not GFP. (D) Bar graph showing the number of GFP‐negative, RFP‐positive spores for strains expressing GFP from chromosome 1, 12 or 14, averaged over two repeats. This number is significantly (P = 1.6 × 10−5) lower for strains with GFP located on chromosome 12.
In this study, we set out to test whether or not certain (core or LS) chromosomes of Fol4287, including chromosome 14, were indeed CD and, if so, what were the phenotypic consequences. By tagging selected chromosomes with a gene for green fluorescent protein (GFP) and screening by flow cytometry for spores without GFP, we were able to estimate the rate of loss for core chromosomes 1 and 12, and LS chromosome 14. Loss of chromosome 1 was not observed. In contrast, strains were obtained in which LS chromosome 14 or, surprisingly, core chromosome 12 sustained large deletions or were even completely absent. Whole‐genome sequencing revealed that some deletions occurred at the same location.
Results
LS chromosomes and a core chromosome can be spontaneously lost during vegetative growth
To assess possible differences in mitotic stability between LS and core chromosomes during vegetative growth, two independent strains were created with an HPH‐GFP fusion gene targeted to a specific chromosome for selection of loss (HPH confers hygromycin resistance). A second fluorescent marker, RFP (red fluorescent protein gene), was randomly integrated into the genome of these two strains to be able to distinguish live cells from cellular debris. The resulting strains with GFP targeted to chromosome 14 are called 14HG1+RFP1, 14HG1+RFP2 and 14HG2+RFP. For the analysis of the stability of a large core chromosome, the HPH‐GFP gene fusion was targeted into a locus approximately 1.4 Mb from one end of chromosome 1 of wild‐type strain Fol4287 (for primers, see Table S1 in Supporting Information). Next, two independent transformants, 1HG1 and 1HG2, were transformed with a plasmid containing RFP, yielding transformants 1HG1+RFP and 1HG2+RFP for use in further experiments.
To determine the GFP loss rate for the large core chromosome 1 and the small LS chromosome 14, the five selected strains were grown for 3 days in minimal medium. Spores were then isolated from the cultures by filtration and screened for the absence or presence of GFP and/or RFP using flow cytometry, as described previously (Vlaardingerbroek et al., 2015). Dot plots were created with the signals from each individual particle (either spore or cellular debris) detected by the GFP and RFP channels. Screening of 106 particles took around 1 min, allowing for high‐throughput screening.
For every tagged chromosome, at least 10 independent cultures were screened for the loss of GFP fluorescence. From each culture, one million spores were analysed and the results were displayed in dot plots. Examples are shown in Fig. 1B, C, E and F. Dot plots showed GFP‐negative, RFP‐positive populations for all of the cultures (Fig. 1B, C, E and F, indicated by arrows). Variation between cultures was high, particularly in cultures of 1HG+GFP strains. Some cultures contained very few spores without GFP, but with RFP present, i.e. live cells potentially missing the region tagged with GFP (Fig. 1C, arrow). Other cultures from the same strains contained more than 1000 GFP‐negative, RFP‐positive spores per million (Fig. 1F, arrow). The differences in the number of RFP‐negative spores may be the result of the different constructs used to insert RFP into the genome.
For chromosome 14, the average number of spores per million lacking GFP was 1175 ± 325 (Fig. 1D). When the gene was located on core chromosome 1, this number was 750 ± 250, which is not significantly lower (P = 0.35).
For each of the 10 independent cultures used per strain, 36 individual GFP‐negative, RFP‐positive spores were deflected to solid medium on which spores were allowed to grow into colonies. Around 15%–20% of the spores gave rise to colonies. For all colonies, the presence or absence of GFP and RFP was checked using fluorescence microscopy. Strains for which loss of fluorescence was confirmed were then tested for the presence of GFP by polymerase chain reaction (PCR). Some strains had lost fluorescence, but retained the gene, which could be the result of the loss of expression or mutations in the gene. Finally, strains lacking GFP were tested for several markers spread over the chromosome (Tabls S2, see Supporting Information). to test for the loss of larger genomic regions. The results for the different chromosomes are summarized in Table 1. In total, we found a loss of (a region of) chromosome 14 in 12 colonies, a loss of (a region of) chromosome 12 in 27 colonies and no colonies that had lost more than 200 kb of chromosome 1.
Table 1.
Selection of strains with loss of (parts of) a chromosome.
| Chromosome with GFP marker | Chr1 | Chr12 | Chr14 |
|---|---|---|---|
| Colonies analysed after flow cytometry | 64 | 103 | 94 |
| GFP‐negative under microscope | 57 | 96 | 82 |
| GFP lost | 36 | 64 | 36 |
| Other markers missing | 0 | 27 | 12 |
To determine which region of chromosome 14 was missing from the 12 strains in which GFP was inserted in chromosome 14, 17 markers spread over chromosome 14 (Fig. 2A) were used to ‘scan’ the chromosome. Six strains were negative for all markers, suggesting loss of the entire chromosome 14; the other six strains had apparently lost part of chromosome 14. As each of the latter six strains lacked the same nine markers (c–l), a similar region of chromosome 14 seemed to be missing (Fig. 2C). The average rate of loss of the entire chromosome 14 was 1 in approximately 65 000 spores. For the large deletions, the rate was an average of 1 in approximately 70 000 spores. Strains that had lost (part of) chromosome 14, which were used for further analysis, were designated as 14‐1–14‐7, where the first number indicates the chromosome that contained the marker. The same convention was used for strains that had lost the marker on chromosome 12.
Figure 2.
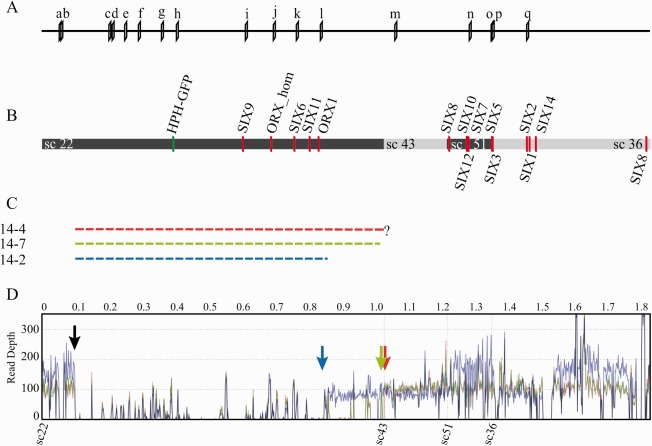
Location of markers and genes encoding proteins secreted in xylem (SIX) on positioned scaffolds of chromosome 14. (A) Location of primer pairs used to determine the size and location of the deletions in chromosome 14. The primer sequences are shown in Table S1 (see Supporting Information). (B) Location of genes for small secreted proteins (red bars) and of the insertion of the GFP (green fluorescent protein gene) marker (green bar) in chromosome 14. Alternating coloured bars indicate different supercontigs. Only positioned supercontigs are displayed. (C) Broken bars indicate the three deletions found, labelled with the strain number and colour coded. The deletion in 14‐4 continues until the end of supercontig 22, but does not continue in supercontig 43; the exact location of the deletion is unknown. All three strains lack the same five genes for in xylem‐secreted proteins. (D) Average read depth of genome sequences mapped to the reference for chromosome 14 for strains 14‐2 (blue line), 14‐7 (green line) and 14‐4 (red line). The black arrow marks the start of the gap in all three strains. The end of each gap is indicated by an arrow in the colour corresponding to the colour of the lines for each sample. Read depth for 14‐2 is higher for part of sc22, all of sc51 and part of sc36.
Contour‐clamped homogeneous electric field (CHEF) gel analysis of four strains (14‐1, 14‐3, 14‐5 and 14‐6) that were negative for all chromosome 14 markers confirmed the loss of chromosome 14 when compared with the wild‐type strain (Fig. 3A). Three strains (14‐2, 14‐4 and 14‐7) that were positive for only some of the chromosome 14 markers contained a new chromosome smaller than chromosome 14 or 15, probably representing the remainder of chromosome 14 (Fig. 3A). Strains 14‐4 and 14‐7 contained a new chromosome of similar size, whereas 14‐2 harboured a larger chromosome (Fig. 3A, indicated by arrows). The absence of chromosome 15 in all of these strains resulted from the unexpected absence of chromosome 15 in the parental strains (14HG1 and 14HG2) (Fig. S1, see Supporting Information). In other experiments, chromosome 15 was also shown to be able to sustain deletions using selection by flow cytometry (Fig. S2, see Supporting Information).
Figure 3.
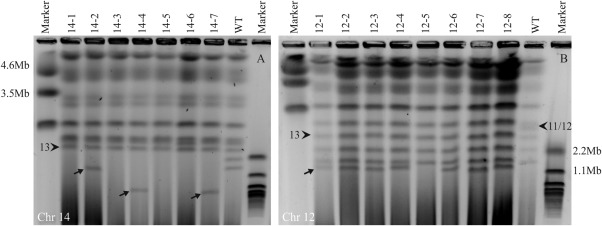
Karyotype analysis confirms partial or complete loss of chromosomes. (A) Contour‐clamped homogeneous electric field (CHEF) gel showing the size and number of chromosomes for strains which have lost chromosome 14 (14‐1, 14‐3, 14‐5 and 14‐6) or a region of chromosome 14 (14‐2, 14‐4 and 14‐7). Chromosome 13 is indicated with an arrowhead for reference. All strains lack the smallest chromosome from the wild‐type (WT) strain (chromosome 15) as well as the normal sized chromosome 14. Strains 14‐2, 14‐4 and 14‐7 have a smaller chromosome absent in the WT strain, indicated with arrows. (B) CHEF gel analysis of strains with partial or complete loss of chromosome 12 (labelled 12‐1–12‐8). Chromosomes 11 and 12, which run closely together, are indicated with an arrowhead. In all the selected strains without GFP (green fluorescent protein gene), the band corresponding to chromosome 12 is missing. Below chromosome 11 in these strains are chromosomes 13–15, in most cases followed by a novel chromosome. This presumed remainder of chromosome 12 is indicated with an arrow in 12‐1. This chromosome is absent in 12‐5, suggesting the complete loss of chromosome 12 in this strain.
Core chromosome 12 is dispensable for in vitro growth
To investigate the stability of a core chromosome with a relatively small size, we tagged chromosome 12 with GFP. Only one transformant was found with the marker inserted at the intended location. This strain, 12HG, was then transformed with a construct containing RFP, yielding 12HG1+RFP1 and 12HG1+RFP2. With an average of 78 ± 17 spores per million lacking GFP (two experiments, each with five cultures per strain), GFP appeared to be relatively stable at this location (Fig. 1E,F). The fraction of spores that grew into a colony was the same as for the other chromosomes tested, around 15%–20%. Of these, 27 were negative for a marker located approximately 450 kb downstream of the site of GFP insertion. A single colony was negative for an additional marker located approximately 1.3 Mb upstream of the site of insertion. The average rate of loss of one of the markers was 1 in 190 000.
On CHEF gels, chromosome 12 migrated close to or together with chromosome 11 (arrowhead in Fig. 3B). Karyotyping of eight strains missing one or both markers of chromosome 12 (strains 12‐1–12‐8) revealed the absence of chromosome 12. Seven of these strains were found to contain a chromosome migrating just below chromosome 15 (indicated by an arrow in Fig. 3B for strain 12‐1). In strain 12‐5, which was negative for both chromosome 12 markers, this small chromosome was not present at all, suggesting the absence of the entire chromosome 12. No obvious growth defects were observed. From this, we conclude that, surprisingly, chromosome 12 is dispensable for in vitro growth of F. oxysporum. The analysis of growth on different carbon sources of this strain is discussed below.
Whole‐genome sequencing reveals deletion hotspots and secondary deletion/duplication events in LS regions
Whole‐genome sequencing was used to determine which regions of chromosomes 12 and 14 were missing in the strains found to contain large deletions, and to confirm the loss of entire chromosomes from other strains. The genomes of three chromosome 12 and three chromosome 14 deletion strains were sequenced, as well as two strains that had probably completely lost either chromosome 12 or 14. Interestingly, mapping of the reads to the reference genome for each of the strains containing a deletion in chromosome 14 revealed that one border of the deletion was in the same region in all three strains; the reads mapped until position 99 132 on supercontig 22. To visualize these deletions, the average number of reads was counted in 10‐kb windows and mapped to the assembly of chromosome 14 (Fig. 2D). In the reference genome, this position was followed by a sequence gap of unknown size, making it impossible to determine the exact location of the start of the deletion. The other border of the deletion was located at a different position in each strain: at position 859 648 in 14‐2, at 1 027 000 in 14‐7 (at both locations, several kb of unique reads mapped to the reference) and between supercontig 22 and supercontig 43 in 14‐4. The estimated sizes of the deletions in 14‐2 and 14‐4 were 760 and 930 kb, respectively. As the distance between supercontig 22 and supercontig 43 was unknown, an estimation for 14‐7 was not possible. Based on the CHEF gel, the size difference between 14‐7 and 14‐4 is minor (Fig. 3A). There appeared to be several regions of chromosome 14 in strain 14‐2 which showed increased read depth compared with the other two samples and 14‐2's own baseline, notably one end of supercontig 22, all of 51 and part of 36. This suggests the presence of segmental duplications and is consistent with the size of this chromosome on the CHEF gel (Fig. 3A), which was much larger than the other two, whereas the size difference of the gap was only 150 kb. No reads were found for which the paired ends mapped to opposite ends of the gap. Attempts to bridge the gaps using primers at either border of the deletions were unsuccessful. Most likely, this is a consequence of repetitive sequences on either end of the deletions.
Mapping the sequencing reads to the reference genome for three strains that had a deletion in chromosome 12 (12‐1, 12‐4 and 12‐8) revealed that the deletions were very similar, even though the strains were isolated from independent cultures (Fig. 4, bottom panel). Curiously, the start of the deletion in all three cases was located at nucleotide 278 722 on supercontig 23 of the reference strain, the exact location of the insertion of the T‐DNA containing GFP. The other border of the deletion was outside supercontig 23 in all three strains, and therefore the exact location could not be identified. In an attempt to find the exact size and location of the deletions in chromosome 12, the sequencing reads were mapped to our own assembly of the genome of strain Fol007, which was very similar to Fol4287. As our own assembly was based on long Pac‐Bio reads, rather than the short reads used for the Fol4287 reference genome, this assembly consisted of much larger contigs (this approach gave no further insights when applied to chromosome 14). Using this approach, we were able to localize the other border of the deletion at nucleotide 155 000 of Fol4287 supercontig 44 in all three strains, an unassigned scaffold in the reference assembly. Examination of this position in the mapping to the Fol4287 reference genome confirmed the position of this border. Consistent with the CHEF gel analysis, the deletions were around 900 kb in length. Located close to this border was a sequence with 70% identity to the dane1 retrotransposon from Aspergillus nidulans. This deletion was flanked by non‐unique sequences. Mapping to our assembly of Fol007 revealed that the start of the sequencing gap was flanked by FOXG_16465, a gene with eight near‐identical homologues in the Fol4287 genome, mostly on LS chromosomes. As with chromosome 14, no reads were found spanning both ends of the deletion. This could be because of the many non‐unique reads present near this end of the deletion.
Figure 4.
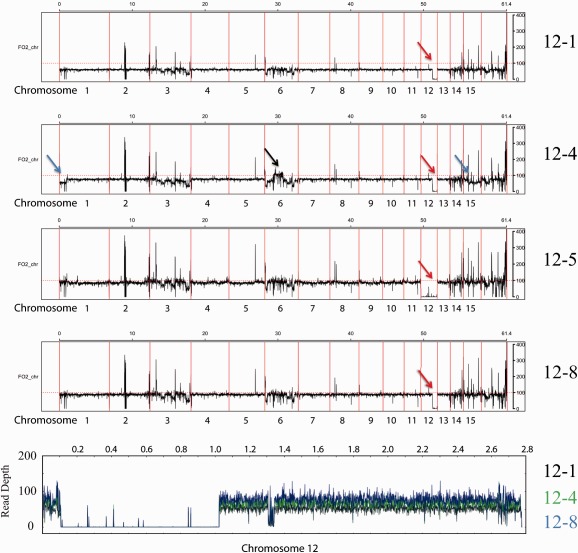
Average read depth reveals duplication and loss events. Average read depth graphs for strains that lost chromosome 12. Strains 12‐1, 12‐4 and 12‐8 all appear to have the same deletion in chromosome 12, indicated by red arrows. The average read depth mapped against the supercontig representing most of chromosome 12 from our own assembly is shown in the bottom panel (12‐1 in black, 12‐4 in green and 12‐8 in blue). Here, we can clearly see that all three strains lack the same region. Strain 12‐4 has a duplicated region on chromosome 6 relative to the reference genome, indicated by a black arrow. This duplication event appears to be very similar to that shown for strain 14‐2 in Fig. 5. Strain 12‐4 has a lower read depth for the lineage‐specific (LS) region of chromosomes 1 and 15 compared with strains 12‐1 and 12‐8, indicated by blue arrows.
To determine whether or not any secondary deletion or duplication events took place on other chromosomes in the sequenced strains, the average read depth was determined for 10‐kb windows and plotted against the reference genome. In general, the average read count was very consistent throughout the genome, with the exception of LS regions, which showed larger variation. These large drops in read depth can be explained by the presence of sequence gaps (Fig. S3, see Supporting Information). This was performed for the three deletions in chromosome 14, for the three deletions in chromosome 12, and for 12‐5 and 14‐1 which, judging from the CHEF gel analysis, seemed to lack the entire chromosome 12 or 14. Comparing 14‐1, 14‐2, 14‐4 and 14‐7 (Fig. 5) with the strains with deletions in chromosome 12 (Fig. 4, panels 12‐1, 12‐4, 12‐5 and 12‐8) revealed that chromosome 14 was indeed completely missing in strain 14‐1, with all read counts well below the baseline of the other chromosomes (Fig. 5, panel 14‐1, green arrow; the deletions in chromosome 14‐2, 14‐4 and 14‐7 are indicated with a green arrow in their respective panels). Consistent with the absence of chromosome 15 in the CHEF gel, the average read depth for chromosome 15 and the start of chromosome 1, which is a duplication of a region of chromosome 15 (Ma et al., 2010), was also lower in the four strains with chromosome 14 deletion or absence. The three strains with the deletion in chromosome 12 showed the exact same region of reduced read depth in chromosome 12 (indicated by red arrows in Fig. 4; the bottom panel displays the entire gap by zooming in using chromosome 12 from the Fol007 assembly as a reference), consistent with our observation that the deletions in the three sequenced strains are the same, and the CHEF gel, although, in the gel, 12‐4 seemed to have a smaller deletion than 12‐1 (Fig. 3B). For strain 12‐5, which appeared to be completely missing chromosome 12 in the CHEF gel, the read depth was close to zero over the full length of the chromosome, confirming loss (Fig. 4, panel 12‐5).
Figure 5.
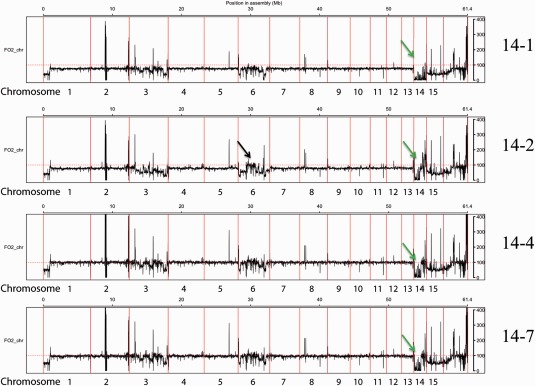
Average read depth reveals duplication and loss events. The panels show the average read depth for the strain from which chromosome 14 was lost completely (14‐1) and three strains which have a 0.9‐Mb deletion in chromosome 14, indicated by green arrows. Strain 14‐2 has a smaller deletion than 14‐4 and 14‐7. Strain 14‐2 has a duplicated region on chromosome 6 relative to the reference genome, indicated by a black arrow.
We also observed that the LS part of the genome had undergone duplications or deletions in several of the strains sequenced. There was variation in the LS region of chromosome 1, which was duplicated on chromosome 15. In strain 12‐4, the read depth for this region was much lower than the baseline for 12‐4, and in 12‐1 and 12‐8, indicating loss of one of these regions relative to the other two strains (Fig. 4, panel 12‐4, blue arrows).
It appeared that there were also duplications of parts of LS chromosome 6 in two strains. Both 14‐2 and 12‐4 showed a region in the middle of the chromosome with a relative read depth clearly higher than that in the other strains (Fig. 5, panel 14‐2, and Fig. 4, panel 12‐4, black arrows). These duplications were independent events because the two strains originated from different parents. The read depth was not doubled as these regions were already present more than once in the genome, and reads that map to more than one location are distributed evenly over all locations to which they map.
As described above, duplications of regions of chromosome 14 also took place in strain 14‐2 (Fig. 2D). Taken together, it is clear that, apart from the events selected for by flow cytometry, LS regions of the genome are highly variable, even between colonies originating from the same culture. These events seem to be similar in location and size, even between different strains and experiments. This is also the case for the deletion events in chromosome 12 and 14. This indicates the presence of duplication and/or deletion hotspots in the F. oxysporum genome.
Loss of chromosome 12 or large portions of chromosome 14 does not significantly affect pathogenicity
Chromosome 14 has been shown to contribute to the infection of tomato when horizontally transferred to a non‐pathogenic strain (Ma et al., 2010). We therefore considered it likely that the loss of chromosome 14 would lead to a reduction in pathogenicity. The effector genes SIX6, SIX9 and SIX11, as well as ORX1 encoding an in xylem‐secreted oxidoreductase and a close homologue of ORX1, are located in the region that is deleted in strains 14‐2, 14‐4 and 14‐7 (Fig. 2). Hence, we expected these strains to also show reduced pathogenicity. No known effector genes are located on chromosome 12, but the expression of many genes located on chromosome 12 is induced on invasion of tomato roots (S. M. Schmidt et al., unpublished results). To assess the contribution to virulence of the entire chromosome and chromosomal regions deleted from chromosomes 12 and 14, a disease assay was performed on 10‐day‐old susceptible tomato seedlings.
The disease index was scored on a scale of 0–4 after 3 weeks, as described previously (Rep et al., 2004). All strains were tested that had partially or completely lost either chromosome 12 or 14. Strains 14HG1+RFP1 (Fig. 1, 6, ‐1) and 14HG1+RFP2 (Fig. 1, 6), from which chromosome 14 deletion strains were derived, also lacked chromosome 15. Loss of the entire chromosome 14 (strains 14‐1, 14‐3, 14‐5 and 14‐6) led to a complete loss of pathogenicity (Fig. 6A,C). To test whether the lack of pathogenicity of strains without chromosome 14 was indeed caused by the absence of chromosome 14 and not other mutations, we re‐introduced chromosome 14 into five strains which had completely lost chromosome 14. This was achieved by transferring chromosome 14 from Fol4287 as described earlier for chromosome 14 of Fol007 (Ma et al., 2010). Although pathogenicity varied between each of the complemented strains, all were clearly pathogenic, confirming that chromosome 14 is required for pathogenicity (Fig. 6D).
Figure 6.

Chromosome 12 and a large region of chromosome 14 are dispensable for pathogenicity. (A) Plants infected with strain 14‐6, which lacks chromosome 14, are completely healthy. (B) Plants infected with strain 14‐7, from which part of chromosome 14 is lost, show disease symptoms similar to those of the wild‐type and parental strains. (C) Distribution of disease index (DI) for strains which have lost part or all of chromosome 14. DI was scored in the range 0–4 based on the number of brown vessels and macroscopic effects, as described previously (Rep et al., 2004). Twenty plants were used per strain. The experiment was performed twice with similar results. 1‐1 indicates 14HG1+RFP1 and 1‐2 indicates 14HG1+RFP2; these are the strains from which the other strains were selected and they show disease severity similar to that of the wild‐type Fol4287 (despite the absence of chromosome 15). 14‐7 was selected from a culture of 14HG1+RFP2, whereas the other strains all originate from 14HG1+RFP1 cultures. The pathogenicity of strains 14‐2, 14‐4 and 14‐7, which have all sustained a deletion in chromosome 14, is not significantly different from that of the wild‐type. Strains completely lacking chromosome 14 have lost pathogenicity. (D) DI for five independent strains (labelled C1–C5) derived from chromosome 14‐lacking strains complemented with chromosome 14 through horizontal chromosome transfer. All strains show a regain of pathogenicity. (E, F) Photographs of plants infected with strains 12‐1 and 12‐4 which have lost part of chromosome 12. (G) DI for strains lacking part or all of chromosome 12. +RFP1 is strain 12HG1+RFP1 from which strains 12‐1–12‐5 originate; strains 12‐6–12‐8 originate from 12HG1+RFP2. (H) Average plant weight for strains that have lost chromosome 14 or sustained large deletions. No significant differences were observed.
Surprisingly, the large deletions in chromosome 14 in strains 14‐2, 14‐4 and 14‐7 had no strong effect on disease severity compared with the wild‐type (Fig. 6B,C; there was a small apparent effect but none of the differences were significant at P < 0.05). This indicates that the in xylem‐secreted proteins encoded within the deletion are dispensable for pathogenicity. When examining plant weight (Fig. 6H), plants infected with strains that had sustained the deletion appeared to be slightly heavier than those infected with the parental strains, although these differences were again not significant.
As chromosome 12 contains many genes that are up‐regulated during infection, loss of this chromosome or regions of it was expected to result in reduced pathogenicity. However, all five strains that sustained a deletion in chromosome 12 (and strain 12‐5 which had lost the entire chromosome) that were derived from cultures of 12HG1+RFP1, as well as the three strains derived from 12HG1+RFP2, showed a very similar distribution of disease severity compared with the control strains (Fig. 6E–G). Clearly, chromosome 12 is dispensable for pathogenicity towards tomato seedlings.
Loss of chromosome 12 or 14 has limited effects on the utilization of diverse carbon sources
To further investigate the phenotypic implications of the loss of chromosome 12 or 14 (strains 12‐5 and 14‐1) or regions thereof (strains 12‐1 and 14‐2), carbon source utilization by the respective strains was assessed. Strains were grown on Biolog FF plates containing 96 different carbon sources and their growth was quantified as described previously (Michielse et al., 2009a). Absorption was measured and used to cluster both the strains tested and the different carbon sources (Fig. S4, see Supporting Information). Although replicates grouped together, differences between strains were very small. The four carbon sources which showed the greatest differences between strains were tested in a plate assay using minimal medium supplemented with 1% of d‐fructose, d‐ribose, AMP or N‐acetyl‐d‐glucosamine. Differential growth could only be confirmed on d‐ribose: strains completely lacking chromosome 12 (12‐5) showed reduced growth on these plates. This could be the result of losing one of its two ribokinases, FOXG_13522, which is lost in 12‐5, but still present in 12‐1, which showed normal growth (Fig. S5, see Supporting Information).
Discussion
It has been well established that there is a large variation in both chromosome number and size between different strains of F. oxysporum (Ma et al., 2010). Here, we provide an insight into the occurrence and frequency of two of the processes underlying this variability: loss and duplication of genomic regions. We show that loss as well as duplication of genetic material occurs during in vitro growth. The frequency of the loss of, or deletions in, LS chromosomes observed was approximately 1 in 35 000 spores. Loss of (a region of) core chromosome 12 was much less frequent, even though this chromosome is not required for normal growth (except on ribose). This difference in frequency could be the result of different physical properties of core and LS chromosomes, such as the density of repetitive elements. It is interesting to note that these numbers are orders of magnitude smaller than for loss during meiosis (Balesdent et al., 2013; Wittenberg et al., 2009)
Genome plasticity in other asexual pathogens has been suggested to play an important role in adaptation to the host niche (Chuma et al., 2011; de Jonge et al., 2013; Seidl and Thomma, 2014). Chromosome transfer and (partial) loss, as well as recombination and duplications, allow the fungus to adapt to changes in host availability and susceptibility by creating genetic variation in the absence of a sexual cycle. Even when not selected for, we observed duplications and deletions within the LS part of the genome in two of the eight strains examined. It is interesting to note that, in both of these cases (strains 12‐4 and 14‐2), more than one chromosome was affected.
What causes the instability of LS chromosomes?
Both chromosomes 12 and 14 are dispensable for growth, but the loss of chromosome 12 was much less frequent: it was only observed once after screening millions of spores. The large core chromosome 1 was even more stable: only small deletions with a size of <200 kb were observed. Clearly, the small core chromosome 12 is mitotically quite stable and conserved during evolution, even though it is dispensable for growth, pathogenicity and the utilization of most carbon sources tested. We assume that it must confer advantages under conditions that we have not explored, such as growth in soil or competition with other microbes. Nevertheless, we suspect that structural differences, and not reduced fitness of the resulting strains, underlie the differences in loss rates between chromosome 12 and chromosomes 14 and 15. Certain chromosomes may also influence each other's stability; the loss of chromosome 15 from the parental lines, for instance, could have negatively influenced the stability of chromosome 14 in our experiments.
Another difference between core and LS chromosomes is the frequent occurrence of spontaneous duplications or deletions of regions in the latter. Similar events have been described in a number of transformants of F. graminearum (Urban et al., 2015). Deletions were more prevalent in, but not entirely restricted to, the regions of local polymorphism described previously (Cuomo et al., 2007). In our experiments, using a limited number of strains, spontaneous deletions and duplications occurred only in LS regions of the genome.
Deletion hotspots
Deletions within a chromosome and chromosomal breaks in Fusarium species appear to be non‐random. Recombination between Helitron transposable elements in chromosome 14 appears to have been frequently responsible for the evolution of race 2 from race 1 in F. oxysporum f. sp. lycopersici (B. Chellappan et al., unpublished results). Kistler et al. (1996) showed that it is possible to induce chromosomal breaks by the introduction of telomeric repeats. The introduction of the fluorescent marker in chromosome 12 could have functioned in a similar way, by promoting recombination with a specific sequence. Whether the loss of chromosome 12 would also occur without the introduction of such a hotspot remains unclear. If the hotspot reduced stability, this would mean that chromosome 12 is even more stable under natural conditions. Why similar effects were not observed in chromosome 14 remains unexplained. Potentially, the ability of an insertion to facilitate chromosomal breaks is position dependent.
In the present study, we could not determine specific sequence elements involved in the deletions in chromosome 14. This could be the result of repetitive DNA making it difficult to map the genome reads to the exact site of the deletions. We encountered the same problem in the case of chromosome 12, even though it has more than 10 times fewer transposable elements (Ma et al., 2010). One end of the deletion was located in an unpositioned scaffold enriched in non‐unique sequences. The other end of the deletion occurred exactly at the site of insertion of the T‐DNA used for selection. Apparently, the insertion of T‐DNA led to enhanced local recombination at this site, in a surprisingly accurate manner.
Phenotypic effects of chromosomal loss or partial deletion
Complete loss of chromosome 12, 14 or 15 had no or limited effects on the growth and development of Fol4287 during in vitro growth. Nevertheless, the conservation of chromosome 12 across Fusarium species and the conservation of chromosome 14 in Fol suggest that there are circumstances in the environment of Fol under which the presence of these chromosomes is beneficial. For chromosome 14, this would obviously include the ability to proliferate within tomato plants. Alternatively, some chromosomes may (partially) promote their own spread in the population without being beneficial, in which case they can be considered as parasitic or ‘selfish’.
Surprisingly, deletion of a large portion of chromosome 14 had only a small effect on pathogenicity in our disease assays (Fig. 2). The region that was deleted contains the genes encoding Six6, Six9, Six11 and Orx1, which are proteins found in the xylem sap of infected tomato plants (Ma et al., 2010; Schmidt et al., 2013). Six6 has been shown previously to be required for full pathogenicity (Gawehns et al., 2014). In the work by Gawehns et al. (2014), no difference in disease index was found when comparing SIX6 deletion strains with wild‐type strains, but there was a significant difference in the weight of infected plants. This effect may be present, but was not statistically significant, in the current study. When comparing the average weight of plants infected with the deletion strains with their parental strains, P values were 0.054, 0.27 and 0.16 (paired, two‐sided t‐test) for 14‐4, 14‐2 and 14‐7, respectively. Our plants were generally smaller than those of Gawehns et al. (2014), possibly causing smaller differences that were not significant. It is clear, in any case, that the strains lacking 0.9 Mb of sequence from chromosome 14 are able to cause disease in tomato plants, indicating that genes in the remaining part of this chromosome are sufficient for tomato infection. Extending the approach employed in this study, strains lacking different regions of chromosome 14 could be created by tagging a different location on the chromosome and screening for additional deletions in this chromosome. In this way, the minimal set of genes required for infection could be determined.
Surprisingly, the differences in carbon source utilization between wild‐type strains and strains lacking either chromosome 12 or 14, or regions thereof, were very small. Apparently, both the LS chromosome 14 and the core chromosome 12 do not encode important (unique) metabolic capabilities under the conditions tested, except growth on ribose, which requires chromosome 12, possibly because of a ribokinase gene on that chromosome.
Conclusions
We report here the first estimate of the rates of loss or partial deletion during in vitro growth for both LS and core chromosomes, processes that explain part of the karyotype variability found in natural populations of F. oxysporum. Core chromosome 12 proved to be more stable than LS chromosome 14, even though this core chromosome is dispensable for pathogenicity and in vitro growth. The LS part of the genome displays high levels of plasticity, including deletion and duplication events in regions that were not selected for. The method described here to select for loss or partial deletions of chromosomes provides a unique tool for the analysis of the function of CD chromosomes. The identification of chromosomal features that contribute to deletion or duplication hotspots would greatly contribute to our understanding of the mechanisms underlying genome plasticity in F. oxysporum.
Experimental Procedures
Fungal strains
The non‐pathogenic strain used as a control in the bioassays was Fo47 (Lemanceau and Alabouvette, 1991). Strains containing the insertion of HPH‐GFP in chromosome 14 have been described previously (Vlaardingerbroek et al., 2015). Fol4287 (Di Pietro and Roncero, 1996) was transformed with p1HG, p12HG and p15HG according to Mullins et al. (2001) to mark each chromosome with HPH‐GFP. Each of these plasmids was obtained by cloning two adjacent ∼1‐kb fragments for a specified location in the respective chromosome in the two multiple cloning sites of pPK2‐HPH‐GFP (Michielse et al., 2009b) (primers listed in Table S1). Strains 1HG1 and 1HG2 were transformed with pPK2‐HPH‐RFP (Van Der Does et al., 2008a, b) using the FAST protocol (Vlaardingerbroek et al., 2015) to create 1HG1GHR and 1HG2GHR. Strains 12HG1, 14HG1 and 14HG2 were transformed with pGRB (Vlaardingerbroek et al., 2015) to create 12HG1GRB1, 12HG1GRB2, 14HG1GRB and 14HG2GRB.
Preparation of spores for sorting
The preparation of samples and sorting were performed as described previously (Vlaardingerbroek et al., 2015).
CHEF gel analysis
The preparation of protoplasts and running of CHEF gels were performed as described previously (Teunissen et al., 2002) with slight adaptations: 50 mg/mL Glucanex (Sigma‐Aldrich Chemie B.V., Zwijndrecht, The Netherlands) and 5 mg/mL driselase (Sigma) in sorbitol solution were used rather than the 5 mg/mL of Novozyme and 100 mg/mL Glucanex used by Teunissen et al. (2002).
Bioassays
Bioassays were performed as described previously (Rep et al., 2004).
Whole‐genome sequencing
DNA was isolated by phenol–chloroform extraction performed on freeze‐dried mycelium of 5‐day‐old cultures grown in liquid medium (0.17% yeast nitrogen base, 10 mm KNO3 and 3% sucrose). Library construction and Illumina Inc. (San Diego, California, USA) sequencing with an average coverage of at least 50× with 125‐bp paired‐end reads were performed by Keygene NV (Wageningen, the Netherlands). Mapping was performed using CLC Genomics Workbench (CLC bio, Aarhus, Denmark). Reads were trimmed (5′ end, 15 nucleotides) to remove adapter sequences and mapped to the F. oxysporum genome assembled in chromosomes or supercontigs [Fusarium Comparative Sequencing Project, Broad Institute of Harvard and MIT (http://www.broadinstitute.org/)] with 90% coverage and 90% identity set as the lower limit. For the location of the deletion in chromosome 12, reads were also mapped to our own assembly of strain Fol007 using the same parameters. For the visualization of read counts, the mapped reads were sorted using the SAMtools sort function. Duplicate reads were removed using Picard Tools with standard settings; 10‐kb non‐overlapping sliding windows are displayed.
Carbon source utilization
Carbon source utilization was determined by growth on 96 carbon sources on Biolog (Hayward, California, USA) FF plates, as described previously (Michielse et al., 2009a). For hierarchical clustering, absorption values were log‐transformed and then clustered using Gene Cluster 3 (de Hoon et al., 2004), and the resulting files were visualized using Java Treeview (Saldanha, 2004).
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Karyotypes of strains used for sorting the loss of chromosomes 12 and 14. Contour‐clamped homogeneous electric field (CHEF) gel analysis reveals that both 14HG1 and 14HG2 are lacking chromosome 15, which is present in both the wild‐type strain and all strains with the marker on chromosome 12. Spontaneous rearrangements are also visible in 14HG2+RFP2. Photograph is made up of two separate gels. WT, wild‐type.
Fig. S2 Karyotypes reveal a deletion in chromosome 15. Contour‐clamped homogeneous electric field (CHEF) gel showing the karyotype of six individual strains which were selected for loss of the green fluorescent protein (GFP) signal. All six originate from the same culture. Chromosome 14 in the selected strains and chromosome 15 in the wild‐type (WT) are indicated with numbered arrows. Chromosome 15 in these strains, indicated with the unnumbered arrow, is clearly reduced in size compared with the WT chromosome 15.
Fig. S3 GC content and sequence gaps. GC content and sequence gaps were calculated in 8000‐bp sliding windows with a slide size of 2000 bp, calculated with bedtool, and plotted using a custom python script and gnuplot. The red line indicates the GC content and the black line indicates the amount of ‘N’ nucleotides (sequence gaps). The distribution of GC content is quite even throughout the genome. Drops in GC content coincide with sequence gaps and areas of low read density in our biological samples (Figs 4 and 5).
Fig. S4 Carbon source utilization of strains with deletions in, or loss of, chromosomes 12 and 14. Hierarchical clustering of absorption measured at 595 nm after growth on 96 different carbon sources for strains lacking parts (strains 12‐1 and 14‐2) or all (strains 12‐5 and 14‐1) of chromosome 12 or 14. Black cells indicate no growth; bright yellow indicates the highest growth (determined by measurement of the absorption at 595 nm) in the assay.
Fig. S5 Growth assay on different carbon sources on plates. (A, B) 12HG+RFP1 (top) and 12‐5 (bottom), which completely lacks chromosome 12, growing on plates with ribose as carbon source. The growth of 12‐5 is clearly impaired relative to 12HG+RFP1. (C, D) 14HG1+RFP1 (top) and 14‐2 (bottom), which has a deletion in chromosome 14, growing on plates with adenosine‐5′‐monophosphate (AMP) as carbon source. The growth of both strains is very similar.
Table S1 Primers used for cloning.
Table S2 Markers for chromosomes.
Acknowledgements
We thank Lotje van der Does for help with the hierarchical clustering and Like Fokkens for providing Figs 2D and 4 (bottom panel). We thank Jiming Li for help with practical work for the resubmission. This work was made possible by a Vici Grant from the Netherlands Organization for Scientific Research (NWO) to MR.
References
- Balesdent, M. , Fudal, I. , Ollivier, B. , Bally, P. , Grandaubert, J. , Eber, F. , Chèvre, A. , Leflon, M. and Rouxel, T. (2013) The dispensable chromosome of Leptosphaeria maculans shelters an effector gene conferring avirulence towards Brassica rapa . New Phytol. 198, 887–898. [DOI] [PubMed] [Google Scholar]
- Chuma, I. , Isobe, C. , Hotta, Y. , Ibaragi, K. , Futamata, N. , Kusaba, M. , Yoshida, K. , Terauchi, R. , Fujita, Y. and Nakayashiki, H. (2011) Multiple translocation of the AVR‐pita effector gene among chromosomes of the rice blast fungus Magnaporthe oryzae and related species. PLoS Pathog. 7, e1002147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, J.J. , Rounsley, S.D. , Rodriguez‐Carres, M. , Kuo, A. , Wasmann, C.C. , Grimwood, J. , Schmutz, J. , Taga, M. , White, G.J. and Zhou, S. (2009) The genome of Nectria haematococca: contribution of supernumerary chromosomes to gene expansion. PLoS Genet. 5, e1000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covert, S.F. (1998) Supernumerary chromosomes in filamentous fungi. Curr. Genet. 33, 311–319. [DOI] [PubMed] [Google Scholar]
- Croll, D. and McDonald, B.A. (2012) The accessory genome as a cradle for adaptive evolution in pathogens. PLoS Pathog. 8, e1002608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuomo, C.A. , Guldener, U. , Xu, J.R. , Trail, F. , Turgeon, B.G. , Di Pietro, A. , Walton, J.D. , Ma, L.J. , Baker, S.E. , Rep, M. , Adam, G. , Antoniw, J. , Baldwin, T. , Calvo, S. , Chang, Y.L. , Decaprio, D. , Gale, L.R. , Gnerre, S. , Goswami, R.S. , Hammond‐Kosack, K. , Harris, L.J. , Hilburn, K. , Kennell, J.C. , Kroken, S. , Magnuson, J.K. , Mannhaupt, G. , Mauceli, E. , Mewes, H.W. , Mitterbauer, R. , Muehlbauer, G. , Munsterkotter, M. , Nelson, D. , O'Donnell, K. , Ouellet, T. , Qi, W. , Quesneville, H. , Roncero, M.I. , Seong, K.Y. , Tetko, I.V. , Urban, M. , Waalwijk, C. , Ward, T.J. , Yao, J. , Birren, B.W. and Kistler, H.C. (2007) The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science, 317, 1400–1402. [DOI] [PubMed] [Google Scholar]
- Dean, R. , van Kan, J.A.L. , Pretorius, Z.A. , Hammond‐Kosack, K.E. , Di Pietro, A. , Spanu, P.D. , Rudd, J.J. , Dickman, M. , Kahmann, R. , Ellis, J. and Foster, G.D. (2012) The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro, A. and Roncero, M.I.G. (1996) Endopolygalacturonase from Fusarium oxysporum f. sp. lycopersici: purification, characterization, and production during infection of tomato plants. Phytopathology, 86, 1324–1330. [Google Scholar]
- Gawehns, F. , Houterman, P.M. , Ichou, F.A. , Michielse, C.B. , Hijdra, M. , Cornelissen, B.J. , Rep, M. and Takken, F.L. (2014) The Fusarium oxysporum effector Six6 contributes to virulence and suppresses I‐2‐mediated cell death. Mol. Plant–Microbe Interact. 27, 336–348. [DOI] [PubMed] [Google Scholar]
- Goodwin, S.B. , M'barek, S.B. , Dhillon, B. , Wittenberg, A.H. , Crane, C.F. , Hane, J.K. , Foster, A.J. , Van der Lee, T.A.J. , Grimwood, J and Aerts, A. (2011) Finished genome of the fungal wheat pathogen Mycosphaerella graminicola reveals dispensome structure, chromosome plasticity, and stealth pathogenesis. PLoS Genet. 7, e1002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta, R. , Ito, K. , Hosaki, Y. , Tanaka, T. , Tanaka, A. , Yamamoto, M. , Akimitsu, K. and Tsuge, T. (2002) A conditionally dispensable chromosome controls host‐specific pathogenicity in the fungal plant pathogen Alternaria alternata . Genetics, 161, 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon, M.J. , Imoto, S. , Nolan, J. and Miyano, S. (2004) Open source clustering software. Bioinformatics, 20, 1453–1454. [DOI] [PubMed] [Google Scholar]
- Hu, J. , Chen, C. , Peever, T. , Dang, H. , Lawrence, C. and Mitchell, T. (2012) Genomic characterization of the conditionally dispensable chromosome in Alternaria arborescens provides evidence for horizontal gene transfer. BMC Genomics, 13, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, L.J. , Johnson, R.D. , Akamatsu, H. , Salamiah, A. , Otani, H. , Kohmoto, K. and Kodama, M. (2001) Spontaneous loss of a conditionally dispensable chromosome from the Alternaria alternata apple pathotype leads to loss of toxin production and pathogenicity. Curr. Genet. 40, 65–72. [DOI] [PubMed] [Google Scholar]
- de Jonge, R. , Bolton, M.D. , Kombrink, A. , van den Berg, G.C. , Yadeta, K.A. and Thomma, B.P. (2013) Extensive chromosomal reshuffling drives evolution of virulence in an asexual pathogen. Genome Res. 23, 1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler, H.C. , Meinhardt, L.W. and Benny, U. (1996) Mutants of Nectria haematococca created by a site‐directed chromosome breakage are greatly reduced in virulence toward pea. Mol. Plant–Microbe Interact. 9, 804–809. [Google Scholar]
- Lemanceau, P. and Alabouvette, C. (1991) Biological control of fusarium diseases by fluorescent Pseudomonas and non‐pathogenic Fusarium. Crop Prot. 10, 279–286. [Google Scholar]
- Ma, L. , van der Does, H.C. , Borkovich, K.A. , Coleman, J.J. , Daboussi, M. , Di Pietro, A. , Dufresne, M. , Freitag, M. , Grabherr, M. , Henrissat, B. , Houterman, P.M. , Kang, S. , Shim, W. , Woloshuk, C. , Xie, X. , Xu, J. , Antoniw, J. , Baker, S.E. , Bluhm, B.H. , Breakspear, A. , Brown, D.W. , Butchko, R.A.E. , Chapman, S. , Coulson, R. , Coutinho, P.M. , Danchin, E.G.J. , Diener, A. , Gale, L.R. , Gardiner, D.M. , Goff, S. , Hammond‐Kosack, K. , Hilburn, K. , Hua‐Van, A. , Jonkers, W. , Kazan, K. , Kodira, C.D. , Koehrsen, M. , Kumar, L. , Lee, Y. , Li, L. , Manners, J.M. , Miranda‐Saavedra, D. , Mukherjee, M. , Park, G. , Park, J. , Park, S. , Proctor, R.H. , Regev, A. , Ruiz‐Roldan, M. , Sain, D. , Sakthikumar, S. , Sykes, S. , Schwartz, D.C. , Turgeon, B.G. , Wapinski, I. , Yoder, O. , Young, S. , Zeng, Q. , Zhou, S. , Galagan, J. , Cuomo, C.A. , Kistler, H.C. and Rep, M. (2010) Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature, 464, 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao, V.P. , Covert, S.F. and VanEtten, H.D. (1991) A fungal gene for antibiotic resistance on a dispensable (“B”) chromosome. Science, 254, 1773–1776. [DOI] [PubMed] [Google Scholar]
- Michielse, C.B. , van Wijk, R. , Reijnen, L. , Manders, E. , Boas, S. , Olivain, C. , Alabouvette, C. and Rep, M. (2009a) The nuclear protein Sge1 of Fusarium oxysporum is required for parasitic growth. PLoS Pathog. 5, e1000637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michielse, C. , van Wijk, R. , Reijnen, L. , Cornelissen, B. and Rep, M. (2009b) Insight into the molecular requirements for pathogenicity of Fusarium oxysporum f. sp. lycopersici through large‐scale insertional mutagenesis. Genome Biol. 10, R4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins, E.D. , Chen, X. , Romaine, P. , Raina, R. , Geiser, D.M. and Kang, S. (2001) Agrobacterium‐mediated transformation of Fusarium oxysporum: an efficient tool for insertional mutagenesis and gene transfer. Phytopathology, 91, 173–180. [DOI] [PubMed] [Google Scholar]
- Raffaele, S. and Kamoun, S. (2012) Genome evolution in filamentous plant pathogens: why bigger can be better. Nat. Rev. Microbiol. 10, 417–430. [DOI] [PubMed] [Google Scholar]
- Rep, M. , Van Der Does, H.C. , Meijer, M. , Van Wijk, R. , Houterman, P.M. , Dekker, H.L. , De Koster, C.G. and Cornelissen, B.J. (2004) A small, cysteine‐rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I‐3‐mediated resistance in tomato. Mol. Microbiol. 53, 1373–1383. [DOI] [PubMed] [Google Scholar]
- Rouxel, T. , Grandaubert, J. , Hane, J.K. , Hoede, C. , van de Wouw, AP. , Couloux, A. , Dominguez, V. , Anthouard, V. , Bally, P. and Bourras, S. (2011) Effector diversification within compartments of the Leptosphaeria maculans genome affected by repeat‐induced point mutations. Nat. Commun. 2, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha, A.J. (2004) Java treeview–extensible visualization of microarray data. Bioinformatics, 20, 3246–3248. [DOI] [PubMed] [Google Scholar]
- Schmidt, S.M. , Houterman, P.M. , Schreiver, I. , Ma, L. , Amyotte, S. , Chellappan, B. , Boeren, S. , Takken, F.L. and Rep, M. (2013) MITEs in the promoters of effector genes allow prediction of novel virulence genes in Fusarium oxysporum . BMC Genomics, 14, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl, M.F. and Thomma, B.P. (2014) Sex or no sex: evolutionary adaptation occurs regardless. Bioessays, 36, 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teunissen, H. , Verkooijen, J. , Cornelissen, B. and Haring, M. (2002) Genetic exchange of avirulence determinants and extensive karyotype rearrangements in parasexual recombinants of Fusarium oxysporum . Mol. Genet. Genomics, 268, 298–310. [DOI] [PubMed] [Google Scholar]
- Urban, M. , King, R. , Hassani‐Pak, K. and Hammond‐Kosack, K.E. (2015) Whole‐genome analysis of Fusarium graminearum insertional mutants identifies virulence associated genes and unmasks untagged chromosomal deletions. BMC Genomics, 16, 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Does, H.C. , Duyvesteijn, R.G.E. , Goltstein, P.M. , van Schie, C.C.N. , Manders, E.M.M. , Cornelissen, B.J.C. and Rep, M. (2008a) Expression of effector gene SIX1 of Fusarium oxysporum requires living plant cells. Fungal Genet. Biol. 45, 1257–1264. [DOI] [PubMed] [Google Scholar]
- Van Der Does, H.C. , Lievens, B. , Claes, L. , Houterman, P.M. , Cornelissen, B.J.C. and Rep, M. (2008b) The presence of a virulence locus discriminates Fusarium oxysporum isolates causing tomato wilt from other isolates. Environ. Microbiol. 10, 1475–1485. [DOI] [PubMed] [Google Scholar]
- VanEtten, H. , Jorgensen, S. , Enkerli, J. and Covert, S.F. (1998) Inducing the loss of conditionally dispensable chromosomes in Nectria haematococca during vegetative growth. Curr. Genet. 33, 299–303. [DOI] [PubMed] [Google Scholar]
- Vlaardingerbroek, I. , Beerens, B. , Shahi, S. and Rep, M. (2015) Fluorescence assisted selection of transformants (FAST): using flow cytometry to select fungal transformants. Fungal Genet. Biol. 76, 104–109. [DOI] [PubMed] [Google Scholar]
- Wang, C. , Skrobek, A. and Butt, T.M. (2003) Concurrence of losing a chromosome and the ability to produce destruxins in a mutant of Metarhizium anisopliae . FEMS Microbiol. Lett. 226, 373–378. [DOI] [PubMed] [Google Scholar]
- Wasmann, C. and VanEtten, H.D. (1996) Transformation‐mediated chromosome loss and disruption of a gene for pisatin demethylase decrease the virulence of Nectria haematococca on pea. Mol. Plant–Microbe Interact. 9, 793–803. [Google Scholar]
- Wittenberg, A.H. , van der Lee, T.A.J. , M'barek, S.B. , Ware, S.B. , Goodwin, S.B. , Kilian, A. , Visser, R.G. , Kema, G.H. and Schouten, H.J. (2009) Meiosis drives extraordinary genome plasticity in the haploid fungal plant pathogen Mycosphaerella graminicola . PLoS One, 4, e5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Karyotypes of strains used for sorting the loss of chromosomes 12 and 14. Contour‐clamped homogeneous electric field (CHEF) gel analysis reveals that both 14HG1 and 14HG2 are lacking chromosome 15, which is present in both the wild‐type strain and all strains with the marker on chromosome 12. Spontaneous rearrangements are also visible in 14HG2+RFP2. Photograph is made up of two separate gels. WT, wild‐type.
Fig. S2 Karyotypes reveal a deletion in chromosome 15. Contour‐clamped homogeneous electric field (CHEF) gel showing the karyotype of six individual strains which were selected for loss of the green fluorescent protein (GFP) signal. All six originate from the same culture. Chromosome 14 in the selected strains and chromosome 15 in the wild‐type (WT) are indicated with numbered arrows. Chromosome 15 in these strains, indicated with the unnumbered arrow, is clearly reduced in size compared with the WT chromosome 15.
Fig. S3 GC content and sequence gaps. GC content and sequence gaps were calculated in 8000‐bp sliding windows with a slide size of 2000 bp, calculated with bedtool, and plotted using a custom python script and gnuplot. The red line indicates the GC content and the black line indicates the amount of ‘N’ nucleotides (sequence gaps). The distribution of GC content is quite even throughout the genome. Drops in GC content coincide with sequence gaps and areas of low read density in our biological samples (Figs 4 and 5).
Fig. S4 Carbon source utilization of strains with deletions in, or loss of, chromosomes 12 and 14. Hierarchical clustering of absorption measured at 595 nm after growth on 96 different carbon sources for strains lacking parts (strains 12‐1 and 14‐2) or all (strains 12‐5 and 14‐1) of chromosome 12 or 14. Black cells indicate no growth; bright yellow indicates the highest growth (determined by measurement of the absorption at 595 nm) in the assay.
Fig. S5 Growth assay on different carbon sources on plates. (A, B) 12HG+RFP1 (top) and 12‐5 (bottom), which completely lacks chromosome 12, growing on plates with ribose as carbon source. The growth of 12‐5 is clearly impaired relative to 12HG+RFP1. (C, D) 14HG1+RFP1 (top) and 14‐2 (bottom), which has a deletion in chromosome 14, growing on plates with adenosine‐5′‐monophosphate (AMP) as carbon source. The growth of both strains is very similar.
Table S1 Primers used for cloning.
Table S2 Markers for chromosomes.


