Figure 2.
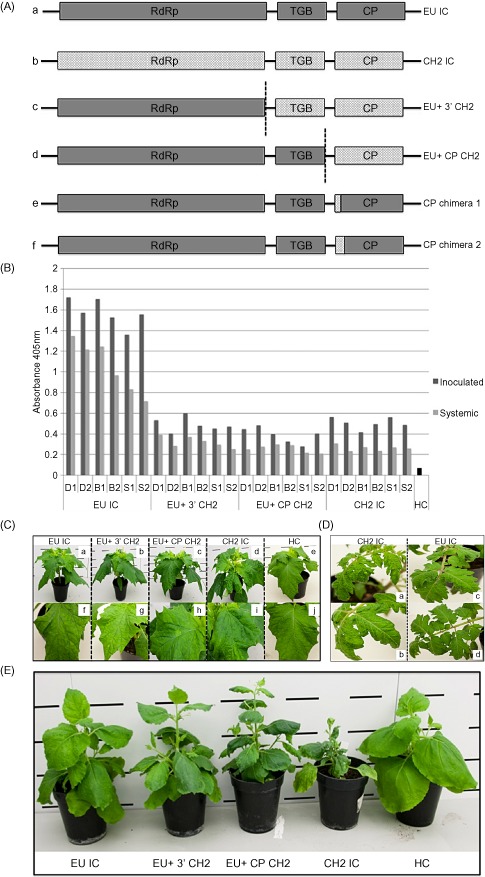
(A) Schematic diagrams of the infectious clones (ICs) generated in this study. Wild‐type (WT) ICs: EU IC (A) and CH2 IC (B). Chimeric ICs: EU+ 3′ CH2 (C), consisting of EU‐derived sequence from the 5′ terminus to the end of the RNA‐dependent RNA polymerase (RdRp) and the remainder CH2 derived to the end of the poly(A) tail; EU+ CP CH2 (D), consisting of EU sequence from the 5′ terminus to the end of TGB3 and the rest CH2 derived to the end of the poly(A) tail; CP chimera 1 and CP chimera 2 (E, F), first 10 and 26 coat protein (CP) amino acids of CH2 origin, respectively, in a mild EU background. (B) Double antibody sandwich‐enzyme‐linked immunosorbent assay (DAS‐ELISA) absorbance data at 21 days post‐inoculation (dpi), displaying local and systemic viral titres (given in dark grey and light grey, respectively) in three species of indicator plant, Datura stramonium (D), Nicotiana benthamiana (B) and Solanum lycopersicum (S), inoculated with EU IC, EU+ 3′ CH2, EU+ CP CH2 and CH2 IC. Healthy (HC) material is included for comparison. (C) Characteristic phenotypes presented by EU IC (a, f), EU+ 3′ CH2 (b, g), EU+ CP CH2 (c, h) and CH2 IC (d, i) in D. stramonium at 21 dpi. Healthy control (HC) plant is given as a comparison (e, j). (D) Characteristic phenotypes presented by EU IC and CH2 IC in S. lycopersicum at 21 dpi. The CH2 isolate displayed a high level of rugosity (a) and vein yellowing (b); EU IC displays an asymptomatic phenotype (c, d). (E) Phenotypes presented by EU IC, EU+ 3′ CH2, EU+ CP CH2 and CH2 IC in N. benthamiana at 21 dpi. A healthy control plant (HC) is given as a comparison.
