Figure 6.
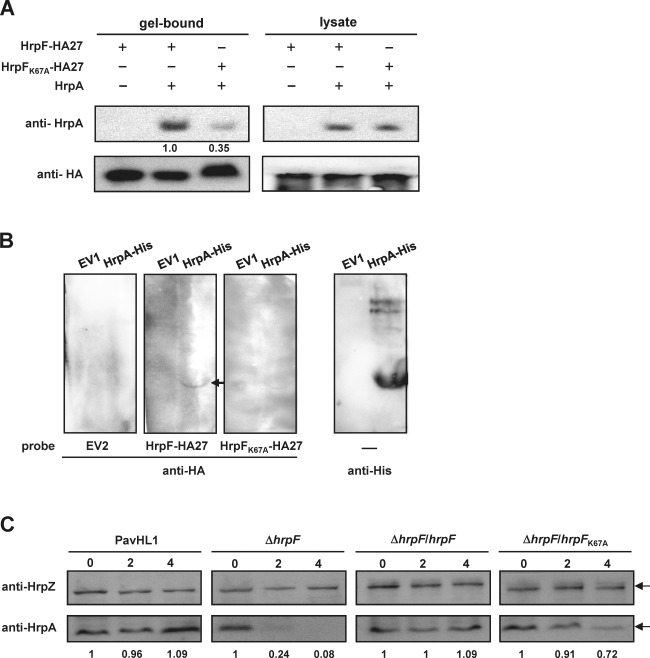
HrpF stabilizes HrpA, and the 67th lysine residue of HrpF is critical for its interaction with HrpA. (A) Immunoblots of immunoprecipitated proteins probed with anti‐HrpA (top) and anti‐haemagglutinin (anti‐HA) (bottom) antibodies to show in vitro protein interactions. Escherichia coli lysates expressing HrpF‐HA27 (pNCHU1867), HrpFK67A‐HA27 (pNCHU1868), HrpA (pNCHU1869) (‘+’) or vector alone (‘−’) were mixed with anti‐HA agarose beads for immunoprecipitation and detected by anti‐HrpA serum as described in Experimental procedures. Gel‐bound, immunoprecipitated proteins; lysate, total proteins as a control to detect the expression of recombinant proteins. (B) Far western analysis shows that HrpF‐HA27 and HrpFK67A‐HA27 bind to HrpA‐His in vitro. Escherichia coli‐expressed HrpA‐His (pNCHU1944) or EV1 (pETite) was probed with bacterial lysates expressing EV2 (pET29a), HrpF‐HA27 (pNCHU1867) or HrpFK67A‐HA27 (pNCHU1868) as described in Experimental procedures. HrpA‐His‐bound HrpF‐HA was further detected with HA monoclonal antibody. (C) Immunodetection of HrpA stability in the Pseudomonas syringae pv. averrhoi (Pav) hrpF mutant. PavHL1, wild‐type harbouring pRK415 and pNCHU2046; ΔhrpF, HL1‐N1589 harbouring pRK415 and pNCHU2046; ΔhrpF/hrpF, HL1‐N1589 harbouring pNCHU1590 and pNCHU2046, ΔhrpF/hrpF K67A, HL1‐N1589 harbouring pNCHU2068 and pNCHU2046. Total cell cultures were harvested at 0, 2 and 4 h post‐chloramphenicol treatment for immunodetection with anti‐HrpZ and anti‐HrpA sera. The stability of HrpA is shown by the fold changes in HrpA banding intensity, which was first normalized by the HrpZ bands and calculated using the value at 0 h post‐chloramphenicol treatment as the denominator.
