Figure 3.
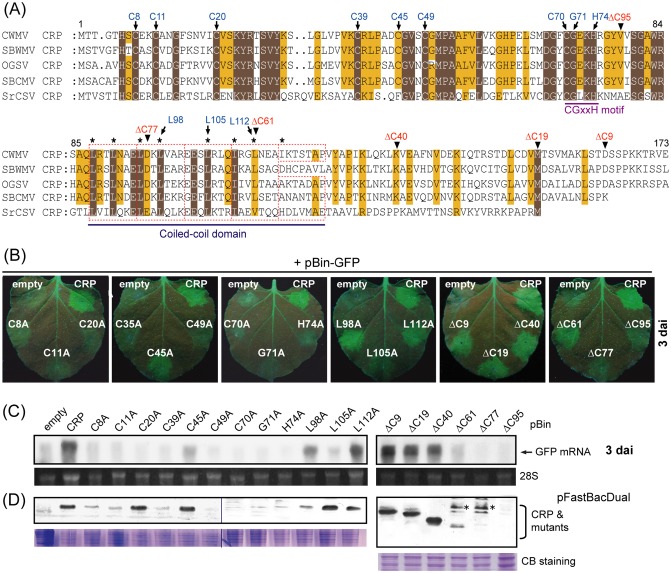
Effects of mutations in conserved amino acid residues and deletion of C‐terminal amino acids on silencing suppression activity and protein stability of Chinese wheat mosaic virus (CWMV) cysteine‐rich protein (CRP). (A) Amino acid sequence alignment of the CRPs encoded by furoviruses. The amino acid positions of CWMV CRP are indicated above the alignment. Dots represent gaps. Dark brown or light brown boxes indicate that amino acids are identical or chemically similar, respectively. Broken red rectangles indicate the predicted heptad repeats. Highly hydrophobic residues at positions a and d of the heptad repeat (abcdefg)n are marked with asterisks. Amino acids that were substituted by alanine are marked with arrows and the C‐terminal ends of the deletion mutants are marked with arrowheads. (B) RNA silencing suppression activity of CWMV mutants in an Agrobacterium co‐infiltration assay. Leaves of Nicotiana benthamiana line 16c plants were infiltrated with mixtures of Agrobacterium harbouring pBin‐GFP plus Agrobacterium harbouring the binary vector containing the CWMV CRP mutants indicated in the images. Green fluorescent protein (GFP) fluorescence was visualized under UV light and photographed at 3 days after inoculation (dai). (C) Northern blot analysis of GFP mRNA accumulation in agroinfiltrated patches of leaves shown in (B). (D) Western blot analysis to detect the accumulation of wild‐type and mutants of CWMV CRP in insect cells. Coomassie blue (CB)‐stained total cell proteins are shown as loading controls (bottom panel). Antibody‐reactive bands of unknown origin are marked with asterisks.
