Figure 1.
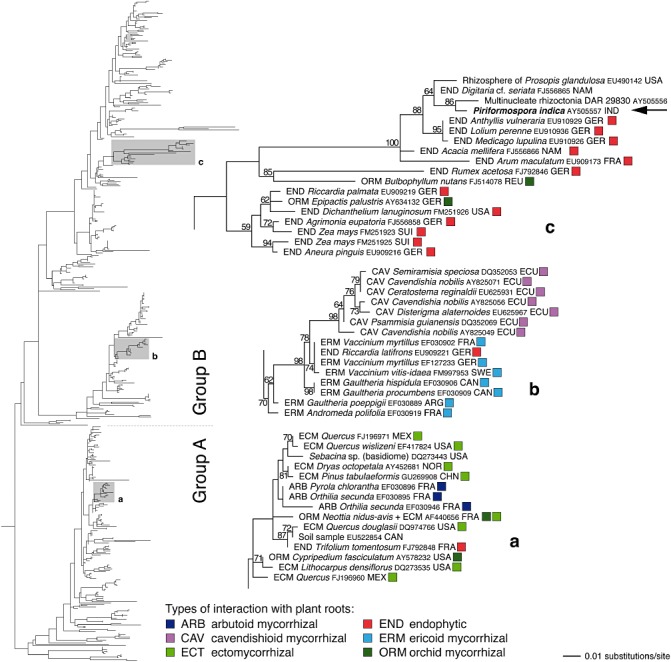
An overview of the phylogenetic relationships in the Sebacinales, based on a representative sampling of partial nuclear‐encoded ribosomal large subunit sequences. Most of the available sequences are from mycobionts detected in plant roots in the field. The alignment was calculated using MAFFT (Katoh and Toh, 2008); a phylogenetic hypothesis was constructed using maximum likelihood analysis as implemented in RAxML (Stamatakis, 2006). Detailed subtrees illustrate the phylogenetic relationships of the three clades highlighted in the full tree (a from Sebacinales group A; b, c from group B); subtree sequences are identified by their GenBank accession numbers. The placement of Piriformospora indica in clade c is indicated by the arrow. Numbers on the branches are bootstrap support values obtained from 1000 replicates (only values ≥50% are shown); branch lengths are scaled in terms of the number of expected substitutions per nucleotide. Coloured boxes indicate the type of symbiosis. Country codes used: ARG, Argentina; CAN, Canada; CHN, China; ECU, Ecuador; FRA, France; GER, Germany; GBR, Great Britain; IND, India; ITA, Italy; MEX, Mexico; NAM, Namibia; NOR, Norway; REU, Reunion; SUI, Switzerland; USA, United States of America. For more details of Sebacinales phylogeny, see Weißet al. (2011).
