SUMMARY
Phytophthora capsici is a highly dynamic and destructive pathogen of vegetables. It attacks all cucurbits, pepper, tomato and eggplant, and, more recently, snap and lima beans. The disease incidence and severity have increased significantly in recent decades and the molecular resources to study this pathogen are growing and now include a reference genome. At the population level, the epidemiology varies according to the geographical location, with populations in South America dominated by clonal reproduction, and populations in the USA and South Africa composed of many unique genotypes in which sexual reproduction is common. Just as the impact of crop loss as a result of P. capsici has increased in recent decades, there has been a similar increase in the development of new tools and resources to study this devastating pathogen. Phytophthora capsici presents an attractive model for understanding broad‐host‐range oomycetes, the impact of sexual recombination in field populations and the basic mechanisms of Phytophthora virulence.
Taxonomy: Kingdom Chromista; Phylum Oomycota; Class Oomycetes; Order Peronosporales; Family Peronosporaceae; Genus Phytophthora; Species capsici.
Disease symptoms: Symptoms vary considerably according to the host, plant part infected and environmental conditions. For example, in dry areas (e.g. southwestern USA and southern France), infection on tomato and bell or chilli pepper is generally on the roots and crown, and the infected plants have a distinctive black/brown lesion visible at the soil line (Fig. 1). In areas in which rainfall is more common (e.g. eastern USA), all parts of the plant are infected, including the roots, crown, foliage and fruit (Fig. 1). Root infections cause damping off in seedlings, whereas, in older plants, it is common to see stunted growth, wilting and, eventually, death. For tomatoes, it is common to see significant adventitious root growth just above an infected tap root, and the stunted plants, although severely compromised, may not die. For many cucurbit fruit, the expanding lesions produce fresh sporangia over days (or even weeks depending on the size of the fruit) and the fruit often look as if they have been dipped in white powdered confectioner's sugar (Fig. 1). Generally, hyphae do not emerge from infected plants or fruit (common with Pythium infections) and all that is visible on the surface of an infected plant is sporangia.
Importance: Phytophthora capsici presents an oomycete worst‐case scenario to growers as it has a broad host range, often produces long‐lived dormant sexual spores, has extensive genotypic diversity and has an explosive asexual disease cycle. It is becoming increasingly apparent that novel control strategies are needed to safeguard food production from P. capsici and other oomycetes. Considering that P. capsici is easy to grow, mate and manipulate in the laboratory and infects many plant species, this pathogen is a robust model for investigations, particularly those related to sexual reproduction, host range and virulence.
Useful websites: Phytophthora capsici genome database: http://genome.jgi‐psf.org/Phyca11/Phyca11.home.html. Molecular tools to identify Phytophthora isolates: http://phytophthora‐id.org.
INTRODUCTION
The filamentous oomycete pathogen Phytophthora capsici causes root, crown, foliar and fruit rot on a number of important vegetables (Erwin and Ribeiro, 1996). Phytophthora capsici was first described in 1922 after it was recovered from chilli pepper at the New Mexico Agricultural Experiment Station field plots in 1918 (Leonian, 1922). It was originally thought to be host specific to pepper, but was soon reported on tomato, eggplant, cucurbits (cucumber, melon, pumpkin and others) and, more recently, green and lima beans (Davidson et al., 2002; Gevens and Hausbeck, 2004; Kreutzer, 1937; Kreutzer and Bryant, 1946; Kreutzer et al., 1940; Tompkins, 1937; Wiant, 1940). Although it is not certain how it is spread over long distances, P. capsici has been reported at locations worldwide, including North and South America, Asia, Africa and Europe (Erwin and Ribeiro, 1996; Hwang and Kim, 1995; Sun et al., 2008). With the emergence of P. capsici on multiple important plant families, efforts have focused on understanding the epidemiology, genetics and mechanisms of infection and virulence (Hausbeck and Lamour, 2004; Quesada‐Ocampo et al., 2011; Ristaino, 1990, 1991; Ristaino and Johnston, 1999). From these studies, it has become clear that, at many locations, P. capsici has a unique life history that routinely employs both sexual outcrossing and rapid asexual reproduction for propagation and survival (Lamour and Kamoun, 2009).
PHYTOPHTHORA CAPSICI TAXONOMY
Recent genus‐wide phylogenetic analyses have revealed that Phytophthora species can be classified into 10 major clades, with P. capsici falling into Clade 2 (Blair et al., 2008) (Fig. 2). A slightly more basal and closely related sister species is P. tropicalis (Aragaki and Uchida, 2001) (Fig. 2). Previously, P. capsici and P. tropicalis were thought to be conspecific on the basis of morphological characters and isozyme data (Mchau and Coffey, 1995; Oudemans et al., 1994). Both species produce deciduous oblong‐shaped sporangia with prominent apical papillae on long pedicels and must outcross to produce amphigynous oospores. Isozyme analysis is not sufficiently sensitive to clearly differentiate between the species and it has only been in the light of DNA sequence data that these species have been more clearly separated. There are a number of general characteristics, in addition to sequence variation, that differ between the two species. Phytophthora capsici isolates are generally recovered from vegetables and do not produce abundant chlamydospores in culture. Phytophthora tropicalis is more often recovered from woody nursery or perennial crops, such as black pepper and cacao. At least one in vitro cross has been successful between P. capsici and P. tropicalis (although many were tried that were unsuccessful), and this led to oospore progeny with alleles from both parents (Donahoo and Lamour, 2008b). In Tennessee, we routinely recover P. tropicalis from nursery‐grown ornamentals, but have never recovered it from nearby vegetable production fields (Donahoo and Lamour, 2008a). The converse is also true, as we have never recovered a P. capsici isolate from the nurseries. There are many reports in the literature that continue to refer to P. tropicalis as P. capsici. This is partly a result of morphological similarities, but also of the deposition of mislabelled internal transcribed spacer (ITS) and other sequences into GenBank.
Figure 2.
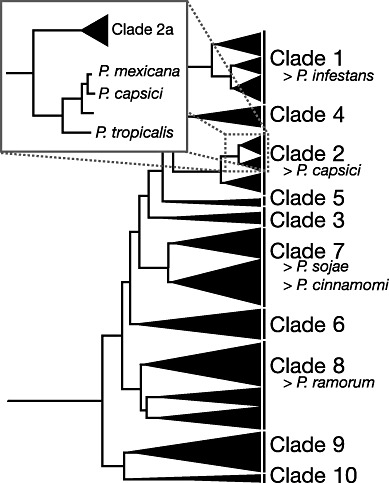
Phylogenetic tree redrawn and modified from Blair et al. (2008). Phytophthora species can be grouped into 10 clades. Phytophthora capsici falls within Clade 2b. Inset, P. capsici clusters with its related sister species P. mexicana and P. tropicalis. Phytophthora species that have or are being sequenced are listed beside the clades.
SEXUAL REPRODUCTION
Phytophthora capsici is a heterothallic species with isolates having one of two mating types (designated A1 and A2). Both mating types are required in close proximity for mating to occur (Ko, 1988). Mating involves a process of stimulation that includes: (i) reception of mating‐type factors (these can be mimicked and are active across a membrane); (ii) transition to the production of both male (antheridia) and female (oogonial) gametangia within each of the A1 and A2 isolates; and (iii) growth of the antheridia through the oogonial gametangia (Erwin and Ribeiro, 1996). Meiosis occurs in the gametangia and haploid nuclei are transported into the oogonium via a fertilization tube. This process occurs within 3–5 days under laboratory conditions and leads to the production of distinctive amphigynous thick‐walled oospores. Oospores formed in the field can persist in the soil for years and are resistant to harsh environmental conditions (Bowers et al., 1990; Lamour and Hausbeck, 2003). Oospores require an indeterminate dormancy period (generally more than 8 weeks) and, depending on the parents, can produce viable sexual progeny when crossed in vitro. It is important to note that (similar to all outcrossing eukaryotes) not all parental combinations are fecund. Crossing is relatively simple, requiring the co‐culture of A1 and A2 mating types on medium that includes a source of sterols (e.g. V8 juice agar) as P. capsici does not make these compounds and they are necessary for viable oospore production (Erwin and Ribeiro, 1996).
Following dormancy, oospores can be stimulated to germinate using a variety of methods, including chemical treatments, passage through live snails or mechanical damage (or a mixture of these) (Erwin and Ribeiro, 1996). An important challenge when isolating oospore‐derived progeny is the separation of the oospores from the subtending mycelium and other asexually produced spores. We routinely use mechanical shearing of the mated culture to separate the oospores from the mycelium, filtration through sterile Kimwipes to separate the oospores from the mycelium and other debris, and overnight treatment with crude enzymes from Trichoderma harzianum to dissolve mycelium and other asexual material and to stimulate oospore germination. Oospores germinate to produce germ tubes that branch into typical mycelium and/or produce sporangia on long pedicels. The sporangia are caducous (deciduous at maturity) and have a prominent papilla (lateral thickening of the sporangial wall).
ASEXUAL REPRODUCTION AND INFECTION
Under favourable environmental conditions, P. capsici will often produce massive numbers of sporangia on the surface of infected tissue (Fig. 1). For cucurbits, this can be particularly pronounced, and the quantification of the number of viable sporangia on the surface of a squash in Michigan revealed that there were no less than three billion viable spores on the surface of this single fruit (K. Lamour, unpublished results). During rain or irrigation events, the mature sporangia are easily dislodged and, when immersed in water, can quickly release 20–40 biflagellate motile zoospores. Zoospores are negatively geotropic (they swim upwards) and will swim chemotactically towards plants. Once at the surface of the plant, the zoospores shed their flagella, encyst and adhere to the surface, and produce a germ tube. The germ tube, aided by secreted enzymes, can penetrate the plant cuticle directly and colonize host tissues (Feng et al., 2010; Li et al., 2011).
Figure 1.
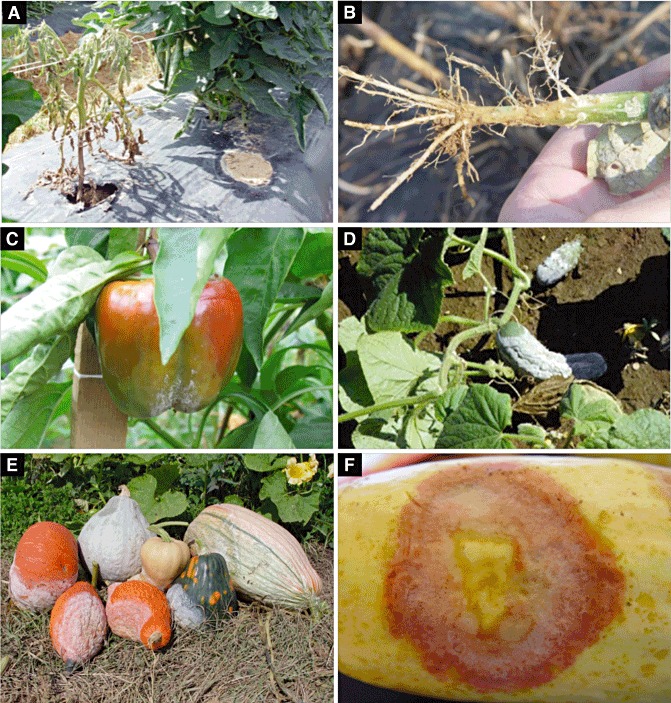
Symptoms associated with Phytophthora capsici infection on various hosts. (A) Tomato plant with root and crown rot caused by P. capsici; note the distinctive brown lesion on the lower part of the plant. (B) Tap root of pumpkin plant showing loss of roots and browning caused by infection. (C) Infected bell pepper with spores on the surface of the fruit. (D) Heavy sporangia production on the surface of infected cucumber fruit. (E) Assortment of ornamental and edible winter squash infected with P. capsici in Tennessee. (F) Natural lesion exhibiting a clearly demarcated island of tissue that contains biotrophic infection surrounded by necrotic tissue with sporangia.
The majority of P. capsici life cycle stages require the presence of a host, as it appears that P. capsici only survives in soil for extended periods as thick‐walled oospores. Infection starts when a hypha emanating from a germinated oospore, a directly germinating sporangium or an encysted zoospore penetrates the plant cuticle and gains access to host cells. Appressoria are observed, in some cases, at the infection site. Successful invasion is then followed by growth and the colonization of host tissues, which ultimately results in tissue collapse and sporulation. Under optimal conditions (25–30 °C and high relative humidity), the time from infection to sporulation occurs within 2–3 days. Consistent with its designation as a hemibiotroph, P. capsici infection features two distinct stages. Infection assays on Nicotiana benthamiana leaf tissues revealed that the early phase features hyphal protrusions (haustoria) that push the host cell membrane inwards and form a direct host–pathogen interface (Schornack et al., 2010) (Fig. 3). Early in infection, cells do not appear to be affected (biotrophy), indicating local suppression of defence responses (E. Huitema, unpublished results) (1, 3). On further ingress, P. capsici switches to necrotrophy, killing infected cells and causing significant tissue collapse and necrosis (1, 3) (E. Huitema, unpublished results). Tissue collapse is then followed by the emergence of sporangia, providing the means for dispersal and a new infection cycle.
Figure 3.
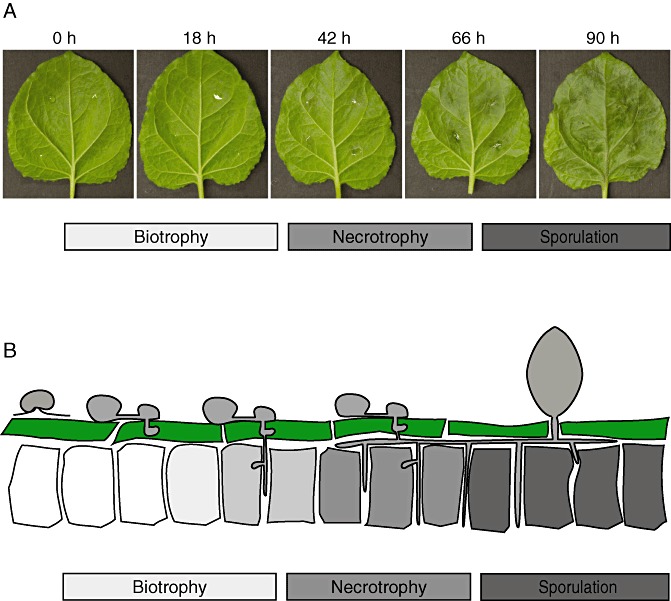
Phytophthora capsici–host interaction studies on the solanaceous model Nicotiana benthamiana. (A) Infection time course assay performed on detached N. benthamiana leaves. Photographs were taken at the time points indicated. Infection features an initial biotrophic phase in which haustoria are observed in colonized tissues. On further ingress (18–42 h), P. capsici lesions become visible and affected tissues collapse. In the later (>66 h) time stages, sporulation ensues and tissue is fully mascerated. (B) Schematic representation of the infection process on N. benthamiana leaves.
The biotrophic phase can lead to significant additional financial losses for P. capsici on fruit. These losses occur because, in many cases, the mature fruit (e.g. cucumber, pumpkin or squash) are infected within a day or two prior to harvest. Because of the lag between the biotrophic and necrotrophic phases, there may be no obvious symptoms when the producer harvests and transports the fruit to a processing station or retailer. After (or on) delivery, the necrotrophic phase of the infection becomes obvious and, in most cases, the processing plant or retailer will reject the fruit. The producer is now responsible for the cost of disposal of the infected produce, and this adds considerable additional losses to those incurred when the disease is obvious in the crop prior to harvest. It also creates a problem of where to (and how to) dispose of the infected and heavily sporulating plant material (Hausbeck and Lamour, 2004).
EPIDEMIOLOGY AND POPULATION BIOLOGY OF P. CAPSICI
Thus far, the epidemiology and population structure of P. capsici have been studied at various locations worldwide, yielding interesting insights into P. capsici population biology. In South Africa and the USA, both mating types are present and populations have a high level of genotypic and genic diversity (presumably as a result of sexual reproduction). The characterization of more than 200 field isolates obtained from pepper production areas along the coast in Peru revealed a single mating type (A2) and the presence of a limited number of genotypes that appeared to represent clonal lineages (Hurtado‐Gonzales et al., 2008). One clonal lineage was clearly dominant, was found across a wide geographical area and persisted in fields across multiple years, suggesting the absence of a sexual cycle. Similar findings have been reported recently for P. capsici infecting pepper in Argentina (Gobena et al., 2011). Here, all the isolates are the A1 mating type. Similar to Peru, a single clonal lineage is spread over a wide area and across multiple years. The findings in Peru and Argentina differ dramatically from reports on P. capsici in the USA and South Africa. In these studies, both mating types were common and the populations had a large proportion of genotypically diverse isolates. At present, there is no conclusive answer to the fundamental differences in P. capsici epidemiology, but it is suspected that year‐round (susceptible) crop production has eliminated the need for sexual reproduction and the formation of oospores (normally needed for survival from season to season). The observation that these clonal isolates are fertile under laboratory conditions appears to support this hypothesis.
RESISTANCE TO P. CAPSICI AND MANAGEMENT
To date, resistance to P. capsici has only been identified in a few select crops. In pepper, studies on the genetics of resistance to P. capsici led to the identification of several major quantitative trait loci (QTLs) (Quesada‐Ocampo and Hausbeck, 2010; Thabuis et al., 2003). Genetic resistance acting against all tested P. capsici isolates, however, is currently only available in the pepper landrace Criollo de Morelos‐CM334, which is not a commercial variety. Without commercial cultivars that harbour durable resistance and because breeding for resistance is complicated by the large number of genetically diverse P. capsici isolates, there is little prospect for resistant commercial pepper cultivars in the near term. In tomato, an extensive screen with four different P. capsici isolates resulted in the identification of one accession, Solanum habrochaites (LA407), that appeared to be resistant to all four strains used (Quesada‐Ocampo and Hausbeck, 2010). Future studies will undoubtedly determine whether the diversity of P. capsici populations will overcome the resistance identified at present in both pepper and tomato. Regardless, an understanding of the molecular mechanisms by which resistance is conferred will be of critical value to growers, breeders and scientists.
Once P. capsici has become established at a location, it can be very difficult to control. Most control strategies aim to limit losses by limiting free water. This is accomplished using a variety of techniques, including planting at well‐drained sites, planting on raised beds, the use of carefully controlled (e.g. trickle) irrigation and, if possible, growing on trellises. Unfortunately, there is no chemical or cultural strategy that is available to limit disease when conditions become warm and wet.
PHYTOPHTHORA CAPSICI REFERENCE GENOME SEQUENCE
Considering the biology and the underlying high level of polymorphisms within P. capsici field populations, a P. capsici genome project was initiated in 2005. The aim was to generate a draft reference genome sequence and to develop a single nucleotide polymorphism (SNP) database. In an effort to reduce the challenges associated with the assembly of a highly polymorphic organism, a series of inbreeding crosses were completed and a moderately inbred isolate was selected for sequencing (Hurtado‐Gonzales and Lamour, 2009). Briefly, an initial cross was completed between two field isolates: an A2 mating type from Tennessee recovered from pumpkin in 2004 (LT263) and an A1 mating type from Michigan recovered from cucumber in 1997 (LT51). Both parents are highly fecund, form dense mats of oospores after mating and produce a high percentage of viable, recombinant oospores. Following this cross, two successive backcrosses to LT263 (the recurrent parent) were made. From the resulting progeny, isolate LT1534 (A2 mating type) was chosen on the basis of a core set of characteristics that included heavy and spontaneous sporulation on simple V8 juice agar, robust oospore production when crossed with (A1) P. capsici isolates and abundant zoospore production from sporangial preparations. Subsequent sequencing of LT1534 with traditional (Sanger) and next‐generation sequencing (NGS) platforms (454 + Illumina) yielded a draft genome sequence covering 64 MB of the predicted 65‐MB (98.4%) P. capsici genome (Phyca11 publicly available at: http://genome.jgi‐psf.org/). The availability of a high‐quality reference genome has enabled in depth studies to be made on various aspects of P. capsici biology, including the mechanisms underpinning infection, genetic diversity and effector evolution.
PHYTOPHTHORA CAPSICI SNP DATA RESOURCES
Analyses with various anonymous molecular markers (amplified fragment length polymorphisms, random amplification of polymorphic DNA and simple sequence repeats) and isozymes have shown that many populations harbour significant genotypic diversity (Dunn et al., 2010; Lamour and Hausbeck, 2001; Meitz et al., 2010; Oudemans and Coffey, 1991). Not surprisingly, a high level of genetic diversity is being uncovered through sequencing projects. Initial estimates based on the re‐sequencing of 20 single‐copy nuclear genes (totalling approximately 11 kb) in a panel of four P. capsici isolates (recovered from Peru, Michigan, Tennessee and New York) revealed a SNP substitution rate of one polymorphic site per 40 bases (K. Lamour, unpublished data). Research is currently underway to identify and validate a large panel of SNP markers for P. capsici using progeny from an in vitro cross. Identification and validation are being accomplished simultaneously using a focused re‐sequencing strategy referred to as restriction site‐associated DNA (RAD) sequencing (Baird et al., 2008). RAD is a cost‐saving strategy that: (i) limits sequencing to the area adjacent to a restriction cut site; and (ii) introduces a molecular tag that allows the pooling of the DNA isolates for sequencing on a next‐generation platform. For P. capsici, genomic DNA was isolated from LT263 and LT51 (parents of the sequenced strain LT1534), as well as from a set of 60 progeny derived from these parents. The DNAs were digested with the restriction enzyme PstI, tagged, pooled and sequenced on a GAII analyser. With approximately 30 000 predicted PstI restriction enzyme sites in the P. capsici genome, RAD sequence data resulted in 10× coverage across 5 Mb of reference sequence between the parents, allowing a broad overview of SNP distribution in the genome (K. Lamour, unpublished results). Preliminary analysis indicated a high level of heterozygosity in the parent isolates with a segregating SNP every 200 bp in these two parents. Validation and mapping of these markers will provide a powerful resource to address a wide variety of important questions, including an assessment of the genes important for pathogenicity and virulence, and reproduction, and will help to identify areas of the genome under positive selection.
INSIGHTS INTO P. CAPSICI EFFECTOR BIOLOGY
The availability of a draft P. capsici genome has accelerated research aiming to address the molecular mechanisms underlying infection. Basic bioinformatic analyses can now be employed to rapidly identify proteins that have been studied extensively in other Phytophthora species, and can now be employed as marker genes in comparative studies. Using such an approach, the initial biotrophic phase, a subsequent switch to necrotrophy and the (onset) of sporulation have been confirmed on various hosts (Fig. 3) (E. Huitema, unpublished results), and further exploration of P. capsici–host associations is underway.
Plant pathogens secrete arsenals of proteins (effectors) that enable parasitic infection and reproduction (Birch et al., 2006; Chisholm et al., 2006; Kamoun, 2006, 2007). Phytophthora species are no different and, in the early stages, infected cells appear to be unaffected and tissues appear to be healthy (biotrophy) (Fig. 3). During the biotrophic phase, a class of effectors, termed RXLRs, is secreted and delivered across the haustorial host–pathogen interface (translocation), where they are thought to aid infection. Secretion and translocation require the presence of a signal peptide, followed by a conserved N‐terminal RXLR motif (Birch et al., 2008; Morgan and Kamoun, 2007; Whisson et al., 2007), features which allow the rapid identification of effector candidates from oomycete (genome) sequences. With the availability of a draft P. capsici genome sequence, genome‐wide searches for RXLR coding genes have identified over 400 putative candidates, pointing to an important role for the RXLR effector type in P. capsici biology (E. Huitema, unpublished results). Subsequent RXLR gene expression analyses on a small subset of candidates have validated this approach and revealed biotrophy‐associated expression in most cases, suggesting a role for these effectors in virulence (E. Huitema, unpublished results). Future studies will reveal the exact roles played by RXLR effectors in P. capsici–host associations.
In addition to the RXLR‐type effectors, another class of cytoplasmic effectors, referred to as ‘crinklers’, has been defined recently in Phytophthora. The Crn1 and Crn2 proteins were first identified in a high‐throughput functional screen of secreted P. infestans proteins and named after their ‘crinkling and necrosis’ phenotypes (CRN) observed on systemic expression in plants (Torto et al., 2003). CRN proteins share a conserved N‐terminal region that harbours a predictable signal peptide (in most cases) and a highly conserved LQLFLAK motif required for translocation. The family features a diverse repertoire of C‐terminal effector domains, which are thought to enhance virulence (Haas et al., 2009; Torto et al., 2003). Computational surveys of the P. capsici genome unveiled approximately 80 full‐length CRN coding gene candidates and more than 200 pseudogenes, suggesting that these effectors are (rapidly) evolving. Further studies on a select set of CRN effector candidates suggested that, similar to their P. infestans family counterparts, the P. capsici effector domain targets the host nucleus, possibly to reprogramme the host and facilitate infection (E. Huitema, unpublished results). To illuminate the host processes or targets modified by P. capsici effectors, studies aimed at dissecting effector function are underway. The identification of effector host targets represents a principal objective of these efforts.
In addition to intracellular effectors, recent studies have implicated other secreted proteins as possible virulence factors. Recently, 18 PcNpp (necrosis‐inducing Phytophthora protein) and nine pectin methylesterase (Pme) coding genes have been identified in P. capsici strain SD33 (Feng et al., 2011; Li et al., 2011). NPP coding genes previously identified in other oomycetes were found to induce host cell death in planta, suggesting a role in host cell perturbation (Fellbrich et al., 2002; Kanneganti et al., 2006; 2002, 2006; Veit et al., 2001). In support of this hypothesis, crystallization studies on the Pythium aphanidermatum NPP‐like protein (NLP) have shown significant structural similarities to cytolytic actinoporins, suggesting cytolytic activity towards pore formation during infection (Ottmann et al., 2009). Considering their high levels of expression, as well as their cell death‐inducing activity, NPP proteins have been proposed to contribute to the transition from biotrophy to necrotrophy. Although 12 PcNpp genes were found to be expressed during infection, their exact functional roles in P. capsici virulence remain to be determined. In addition to the NPP proteins, PcPme genes were also found to be expressed during infection, and exposure of plant tissue to PcPME resulted in tissue collapse and cell death. It is conceivable that their mechanisms of cell death induction and, consequently, their means of contributing to virulence are distinct. Further functional studies should illuminate the roles played by these proteins in the virulence of P. capsici (Li et al., 2011).
P. CAPSICI IN THE POST‐GENOME AGE: A MODEL PATHOGEN OF SCIENTIFIC, STRATEGIC AND ECONOMIC IMPORTANCE
Phytophthora capsici is a pathogen of considerable economic and scientific importance. Because it attacks a broad range of vegetable crops, often in areas in which there has been little research on this pathogen, the overall economic impact is difficult to estimate across crops and continents. In 2010, 77 850 acres of chilli and bell pepper were planted in the USA with an estimated value of $772 million. With P. capsici epidemics causing damage on other major crop hosts (tomato, eggplant, all cucurbits, snap and lima beans) and in other major crop‐growing regions (Central and South America, southern Europe and Asia), worldwide vegetable production valued at well over $1 billion is threatened each year.
In the USA, P. capsici is an introduced pathogen, and movement to a new site will often make it impossible to farm the land that has been used for generations to produce vegetables. Sometimes the location itself is more valuable than the vegetables. For example, in the USA, pumpkins and various winter squash are used extensively for ornamentation during the autumn holidays (e.g. Halloween and various harvest festivals) (Fig. 1). An important component of this autumn business is farms in which consumers can walk around a field and harvest their own pumpkins and squash. In situations in which land for vegetable production is limited (e.g. pumpkin producers on Long Island, NY, USA), pumpkin producers with a history of P. capsici infestation will go so far as to ship in healthy fruit from a distant location to set in their field for consumers to ‘pick their own pumpkins’.
If we consider the absence of resistant cultivars for all crop systems affected and the high level of genotypic diversity and sexual recombination in many field situations, P. capsici presents an oomycete worst‐case scenario for crop production. Knowledge gleaned from this dynamic parasite, particularly knowledge that can help to advance the development of novel control methods, will provide valuable insight into the management of other oomycetes.
Phytophthora capsici has many of the features that could prove to be instrumental in understanding basic Phytophthora biology, particularly for outcrossing species. An ideal model oomycete should be easy to work with, grow rapidly on simple media, sporulate abundantly in culture and be capable of completing crosses that result in viable oospore progeny. The genus Phytophthora is (roughly) split into two groups: those that are self‐fertile and produce oospores in single culture, and those that require outcrossing to produce oospores. The soybean pathogen P. sojae has been proven to be a good model for homothallic species, particularly those that infect a single host. Although outcrossing is possible with P. sojae, recovering outbred versus inbred oospore progeny can be challenging. For outcrossing Phytophthora species, many are difficult (or impossible) to cross in the laboratory. For example, although a reference genome is available for the broad‐host‐range sudden oak death pathogen P. ramorum, in vitro crosses have not been successful. Making things even more difficult is the fact that many outcrossing species, including P. ramorum, produce abundant thick‐walled asexual chlamydospores able to withstand the relatively harsh treatments that allow the isolation of sexual oospores from the subtending mycelium.
Another important characteristic within Phytophthora is between species that infect a limited number of specific hosts and species that infect a broad range of hosts. Phytophthora capsici has an extensive host range that spans several plant families, including the tractable model N. benthamiana, as well as the well‐studied crop plant species tomato and pepper. Genome resources will soon be available for tomato, which is attacked by P. capsici, P. infestans and P. nicotianae, and comparative functional genomics and proteomics investigations will be greatly facilitated. In particular, studies aiming to compare the gene complements required for infection (e.g. effectors) and pathogen perception and defence responses should help to identify conserved versus species‐specific processes.
As the molecular toolbox for the oomycetes continues to grow, the limited success with genetic transformation perhaps remains the last major roadblock in Phytophthora molecular studies. Homologous recombination in Phytophthora is yet to be achieved and heterologous recombination and DNA integration commence at a relatively low rate, limiting the scope for large‐scale in vivo functional analyses. A recently developed P. capsici zoospore electroporation protocol can yield up to 40 transformants per reaction, allowing the rapid and efficient generation of genetically modified laboratory strains (Huitema et al., 2011). As a result of the prolific regeneration and growth rate, transformants can be analysed in a matter of weeks. Using this approach, two novel translocation motifs have been identified recently (Kemen et al., 2011; Schornack et al., 2010). Phytophthora capsici is virulent on plants carrying the R3a resistance gene unless PiAvr3a is expressed in transgenic strains. AVR3a‐triggered immunity requires its delivery inside host cells, a feature that has been exploited by replacing the PiAVR3a RXLR region with candidate translocation signals and testing translocation in vivo (Kemen et al., 2011;Schornack et al., 2010) (Fig. 4). These studies are bringing P. capsici to the fore as a model oomycete pathogen, and we expect the rapid expansion of the oomycete molecular toolbox to continue.
Figure 4.
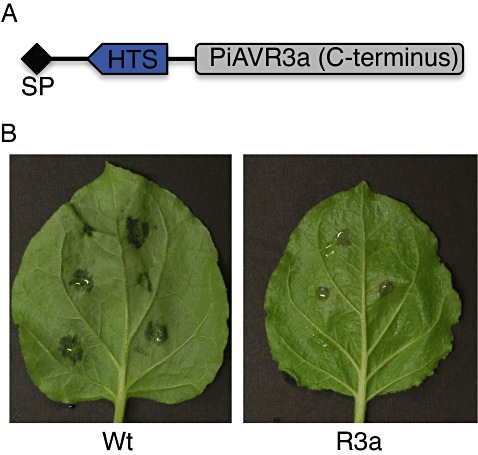
Phytophthora capsici as a model towards an understanding of effector host translocation. Phytophthora capsici transformation allows the development of in vivo host translocation reporters. (A) The PiAVR3a‐based translocation reporter construct has an N‐terminal effector region with a signal peptide (SP), a (conserved) host translocation signal (HTS) and an AVR3a C‐terminal effector domain. (B) Translocation mediated by the host translocation signal is evidenced by the recognition of AVR3a by R3a in transgenic Nicotiana benthamiana leaves. Strains expressing the reporter constructs are virulent on plants lacking R3a (wild‐type, Wt; left), whereas recognition takes place in plants carrying R3a (right).
CONCLUSIONS
Just as the impact of crop loss caused by P. capsici has increased in recent decades, there has been a similar increase in the development of new tools and resources to study this devastating pathogen. The tractability of P. capsici, a reference genome and an unprecedented abundance of molecular variation provide a powerful platform to explore many of the most challenging aspects of Phytophthora parasitism and evolution. In particular, P. capsici presents an attractive model for understanding broad‐host‐range oomycete pathogens, the impact of sexual recombination in field populations, the evolution of clonal lineages and the basic mechanisms of Phytophthora pathogenicity.
ACKNOWLEDGEMENTS
The authors thank Dr Andrew Howden and the anonymous reviewers for their suggestions. This work was supported, in part, by the Royal Society of Edinburgh (co‐funded by Marie‐Curie) and the Biotechnology and Biological Sciences Research Council (BBSRC).
REFERENCES
- Aragaki, M. and Uchida, J. (2001) Morphological distinctions between Phytophthora capsici and P. tropicalis sp. nov. Mycologia, 93, 137–145. [Google Scholar]
- Baird, N.A. , Etter, P.D. , Atwood, T.S. , Currey, M.C. , Shiver, A.L. , Lewis, Z.A. , Selker, E.U. , Cresko, W.A. and Johnson, E.A. (2008) Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE, 3, e3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch, P.R. , Rehmany, A.P. , Pritchard, L. , Kamoun, S. and Beynon, J.L. (2006) Trafficking arms: oomycete effectors enter host plant cells. Trends Microbiol. 14, 8–11. [DOI] [PubMed] [Google Scholar]
- Birch, P.R. , Boevink, P.C. , Gilroy, E.M. , Hein, I. , Pritchard, L. and Whisson, S.C. (2008) Oomycete RXLR effectors: delivery, functional redundancy and durable disease resistance. Curr. Opin. Plant Biol. 11, 373–379. [DOI] [PubMed] [Google Scholar]
- Blair, J.E. , Coffey, M.D. , Park, S.Y. , Geiser, D.M. and Kang, S. (2008) A multi‐locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genet. Biol. 45, 266–277. [DOI] [PubMed] [Google Scholar]
- Bowers, J.H. , Papavizas, G.C. and Johnston, S.A. (1990) Effect of soil temperature and soil‐water matric potential on the survival of Phytophthora capsici in natural soil. Plant Dis. 74, 771–778. [Google Scholar]
- Chisholm, S.T. , Coaker, G. , Day, B. and Staskawicz, B.J. (2006) Host–microbe interactions: shaping the evolution of the plant immune response. Cell, 124, 803–814. [DOI] [PubMed] [Google Scholar]
- Davidson, C.R. , Carroll, R.B. , Evans, T.A. and Mulrooney, R.P. (2002) First report of Phytophthora capsici infecting lima bean (Phaseolus lunatus) in the Mid‐Atlantic Region. Plant Dis. 86, 1049. [DOI] [PubMed] [Google Scholar]
- Donahoo, R.S. and Lamour, K. (2008a) Characterization of Phytophthora species from leaves of nursery woody ornamentals in Tennessee. Hortscience, 43, 1833–1837. [Google Scholar]
- Donahoo, R.S. and Lamour, K. (2008b) Interspecific hybridization and apomixis between Phytophthora capsici and Phytophthora tropicalis . Mycologia, 100, 911–920. [DOI] [PubMed] [Google Scholar]
- Dunn, A.R. , Milgroom, M.G. , Meitz, J.C. , McLeod, A. , Fry, W.E. , McGrath, M.T. , Dillard, H.R. and Smart, C.D. (2010) Population structure and resistance to mefenoxam of Phytophthora capsici in New York State. Plant Dis. 94, 1461–1468. [DOI] [PubMed] [Google Scholar]
- Erwin, D.C. and Ribeiro, O.K. (1996) Phytophthora Diseases Worldwide. St. Paul, MN: The American Phytopathological Society. [Google Scholar]
- Fellbrich, G. , Romanski, A. , Varet, A. , Blume, B. , Brunner, F. , Engelhardt, S. , Felix, G. , Kemmerling, B. , Krzymowska, M. and Nürnberger, T. (2002) NPP1, a Phytophthora‐associated trigger of plant defense in parsley and Arabidopsis. Plant J. 32, 375–390. [DOI] [PubMed] [Google Scholar]
- Feng, B. , Li, P. , Wang, H. and Zhang, X. (2010) Functional analysis of pcpme6 from oomycete plant pathogen Phytophthora capsici . Microb. Pathog. 49, 23–31. [DOI] [PubMed] [Google Scholar]
- Feng, B.Z. , Li, P.Q. , Fu, L. , Sun, B.B. and Zhang, X.G. (2011) Methodology identification of 18 genes encoding necrosis‐inducing proteins from the plant pathogen Phytophthora capsici (Pythiaceae: Oomycetes). Genet. Mol. Res. 10, 910–922. [DOI] [PubMed] [Google Scholar]
- Gevens, A. and Hausbeck, M.K. (2004) Phytophthora capsici isolated from snap bean is pathogenic to cucumber fruit and soybean APS North Central Division Meeting. [Google Scholar]
- Gobena, D. , Roig, J. , Galmarini, C. , Hulvey, J. and Lamour, K. (2011) Genetic diversity of Phytophthora capsici isolates from pepper and pumpkin in Argentina. Mycologia, DOI: 10.3852/11‐147. [DOI] [PubMed] [Google Scholar]
- Haas, B.J. , Kamoun, S. , Zody, M.C. , Jiang, R.H.Y. , Handsaker, R.E. , Cano, L.M. , Grabherr, M. , Kodira, C.D. , Raffaele, S. , Torto‐Alalibo, T. , Bozkurt, T.O. , Ah‐Fong, A.M. , Alvarado, L. , Anderson, V.L. , Armstrong, M.R. , Avrova, A. , Baxter L., Beynon J., Boevink, P.C. , Bollmann, S.R. , Bos, J.I. , Bulone, V. , Cai, G. , Cakir, C. , Carrington, J.C. , Chawner, M. , Conti, L. , Costanzo, S. , Ewan, R. , Fahlgren, N. , Fischbach, M.A. , Fugelstad, J. , Gilroy, E.M. , Gnerre, S. , Green, P.J. , Grenville‐Briggs, L.J. , Griffith, J. , Grünwald, N.J. , Horn, K. , Horner, N.R. , Hu, C.H. , Huitema, E. , Jeong, D.H. , Jones, A.M. , Jones, J.D. , Jones, R.W. , Karlsson, E.K. , Kunjeti, S.G. , Lamour, K. , Liu, Z. , Ma, L. , Maclean, D. , Chibucos, M.C. , McDonald, H. , McWalters, J. , Meijer, H.J. , Morgan, W. , Morris, P.F. , Munro, C.A. , O’Neill, K. , Ospina‐Giraldo, M. , Pinzón, A. , Pritchard, L. , Ramsahoye, B. , Ren, Q. , Restrepo, S. , Roy, S. , Sadanandom, A. , Savidor, A. , Schornack, S. , Schwartz, D.C. , Schumann, U.D. , Schwessinger, B. , Seyer, L. , Sharpe, T. , Silvar, C. , Song, J. , Studholme, D.J. , Sykes, S. , Thines, M. , van de Vondervoort, P.J. , Phuntumart, V. , Wawra, S. , Weide, R. , Win, J. , Young, C. , Zhou, S. , Fry, W. , Meyers, B.C. , van West, P. , Ristaino, J. , Govers, F. , Birch, P.R. , Whisson, S.C. , Judelson, H.S. and Nusbaum, C. (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans . Nature, 461, 393–398. [DOI] [PubMed] [Google Scholar]
- Hausbeck, M.K. and Lamour, K.H. (2004) Phytophthora capsici on vegetable crops: research progress and management challenges. Plant Dis. 12, 1292–1303. [DOI] [PubMed] [Google Scholar]
- Huitema, E. , Smoker, M. and Kamoun, S. (2011) A straightforward protocol for electro‐transformation of Phytophthora capsici zoospores. Methods Mol. Biol. 712, 129–135. [DOI] [PubMed] [Google Scholar]
- Hurtado‐Gonzales, O.P. and Lamour, K.H. (2009) Evidence for inbreeding and apomixis in close crosses of Phytophthora capsici . Plant Pathol. 58, 715–722. [Google Scholar]
- Hurtado‐Gonzales, O. , Aragon‐Caballero, L. , Apaza‐Tapia, W. , Donahoo, R. and Lamour, K. (2008) Survival and spread of Phytophthora capsici in coastal Peru. Phytopathology, 98, 688–694. [DOI] [PubMed] [Google Scholar]
- Hwang, B.K. and Kim, C.H. (1995) Phytophthora blight of pepper and its control in Korea. Plant Dis. 79, 221–227. [Google Scholar]
- Kamoun, S. (2006) A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44, 41–60. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. (2007) Groovy times: filamentous pathogen effectors revealed. Curr. Opin. Plant Biol. 10, 358–365. [DOI] [PubMed] [Google Scholar]
- Kanneganti, T.D. , Huitema, E. , Cakir, C. and Kamoun, S. (2006) Synergistic interactions of the plant cell death pathways induced by Phytophthora infestans Nep1‐like protein PiNPP1.1 and INF1 elicitin. Mol. Plant–Microbe Interact. 19, 854–863. [DOI] [PubMed] [Google Scholar]
- Kemen, E. , Gardiner, A. , Schultz‐Larsen, T. , Kemen, A.C. , Balmuth, A.L. , Robert‐Seilaniantz, A. , Bailey, K. , Holub, E. , Studholme, D.J. , Maclean, D. and Jones, J.D. (2011) Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana . PLoS Biol. 9, e1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko, W. (1988) Hormonal heterothallism and homothallism in Phytophthora . Annu. Rev. Phytopathol. 26, 57–73. [Google Scholar]
- Kreutzer, W.A. (1937) A Phytophthora rot of cucumber fruit. Phytopathology, 27, 955. [Google Scholar]
- Kreutzer, W.A. and Bryant, L.R. (1946) Certain aspects of the epiphytology and control of tomato fruit rot caused by Phytophthora capsici Leonian. Phytopathology, 36, 329–339. [Google Scholar]
- Kreutzer, W.A. , Bodine, E.W. and Durrell, L.W. (1940) Cucurbit diseases and rot of tomato fruit caused by Phytophthora capsici . Phytopathology, 30, 972–976. [Google Scholar]
- Lamour, K. and Kamoun, S. (2009) Oomycete Genetics and Genomics: Diversity, Interactions and Research Tools. Hoboken, NJ: John Wiley & Sons. [Google Scholar]
- Lamour, K.H. and Hausbeck, M.K. (2001) Investigating the spatiotemporal genetic structure of Phytophthora capsici in Michigan. Phytopathology, 91, 973–980. [DOI] [PubMed] [Google Scholar]
- Lamour, K.H. and Hausbeck, M.K. (2003) Effect of crop rotation on the survival of Phytophthora capsici and sensitivity to mefenoxam. Plant Dis. 87, 841–845. [DOI] [PubMed] [Google Scholar]
- Leonian, L.H. (1922) Stem and fruit blight of peppers caused by Phytophthora capsici sp. nov . Phytopathology, 12, 401–408. [Google Scholar]
- Li, P. , Feng, B. , Wang, H. , Tooley, P.W. and Zhang, X. (2011) Isolation of nine Phytophthora capsici pectin methylesterase genes which are differentially expressed in various plant species. J. Basic Microbiol. 51, 61–70. [DOI] [PubMed] [Google Scholar]
- Mchau, G.R.A. and Coffey, M.D. (1995) Evidence for the existence of two subpopulations in Phytophthora capsici and a redescription of the species. Mycol. Res. 99, 89–102. [Google Scholar]
- Meitz, J.C. , Linde, C.C. , Thompson, A. , Langenhoven, S. and McLeod, A. (2010) Phytophthora capsici on vegetable hosts in South Africa: distribution, host range and genetic diversity. Aust. Plant Pathol. 39, 431–439. [Google Scholar]
- Morgan, W. and Kamoun, S. (2007) RXLR effectors of plant pathogenic oomycetes. Curr. Opin. Microbiol. 10, 332–338. [DOI] [PubMed] [Google Scholar]
- Ottmann, C. , Luberacki, B. , Kufner, I. , Koch, W. , Brunner, F. , Weyand, M. , Mattinen, L. , Pirhonen, M. , Anderluh, G. , Seitz, H.U. , Nürnberger, T. and Oecking, C. (2009) A common toxin fold mediates microbial attack and plant defense. Proc. Natl. Acad. Sci. USA, 106, 10 359–10 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudemans, P. and Coffey, M.D. (1991) Isozyme comparison within and among worldwide sources of three morphologically distinct species of Phytophthora . Mycol. Res. 95, 19–30. [Google Scholar]
- Oudemans, P. , Forster, H. and Coffey, M.D. (1994) Evidence for distinct isozyme subgroups within Phytophthora citricola and close relationships with P. capsici and P. citrophthora . Mycol. Res. 98, 189–199. [Google Scholar]
- Quesada‐Ocampo, L.M. and Hausbeck, M.K. (2010) Resistance in tomato and wild relatives to crown and root rot caused by Phytophthora capsici . Phytopathology, 100, 619–627. [DOI] [PubMed] [Google Scholar]
- Quesada‐Ocampo, L.M. , Granke, L.L. , Mercier, M.R. , Olsen, J. and Hausbeck, M.K. (2011) Investigating the genetic structure of Phytophthora capsici populations. Phytopathology, 101, 1061–1073. [DOI] [PubMed] [Google Scholar]
- Qutob, D. , Kamoun, S. and Gijzen, M. (2002) Expression of a Phytophthora sojae necrosis‐inducing protein occurs during transition from biotrophy to necrotrophy. Plant J. 32, 361–373. [DOI] [PubMed] [Google Scholar]
- Qutob, D. , Kemmerling, B. , Brunner, F. , Kufner, I. , Engelhardt, S. , Gust, A.A. , Luberacki, B. , Seitz, H.U. , Stahl, D. , Rauhut, T. , Glawischnig, E. , Schween, G. , Lacombe, B. , Watanabe, N. , Lam, E. , Schlichting, R. , Scheel, D. , Nau, K. , Dodt, G. , Hubert, D. , Gijzen, M. and Nürnberger, T. (2006) Phytotoxicity and innate immune responses induced by NEP1‐like proteins. Plant Cell, 18, 3721–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristaino, J.B. (1990) Intraspecific variation among isolates of Phytophthora capsici from pepper and cucurbit fields in North Carolina. Phytopathology, 80, 1253–1259. [Google Scholar]
- Ristaino, J.B. (1991) Influence of rainfall, drip irrigation, and inoculum density on the development of Phytophthora root and crown rot epidemics and yield in bell pepper. Phytopathology, 81, 922–929. [Google Scholar]
- Ristaino, J.B. and Johnston, S.A. (1999) Ecologically based approaches to management of Phytophthora blight on bell pepper. Plant Dis. 83, 1080–1089. [DOI] [PubMed] [Google Scholar]
- Schornack, S. , Van Damme, M. , Bozkurt, T.O. , Cano, L.M. , Smoker, M. , Thines, M. , Gaulin, E. , Kamoun, S. and Huitema, E. (2010) Ancient class of translocated oomycete effectors targets the host nucleus. Proc. Natl. Acad. Sci. USA, 107, 17 421–17 426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, W.X. , Jia, Y.J. , O'Neill, N.R. , Feng, B.Z. and Zhang, X.G. (2008) Genetic diversity in Phytophthora capsici from eastern China. Can. J. Plant Pathol. 30, 414–424. [Google Scholar]
- Thabuis, A. , Palloix, A. , Pflieger, S. , Daubeze, A.M. , Caranta, C. and Lefebvre, V. (2003) Comparative mapping of Phytophthora resistance loci in pepper germplasm: evidence for conserved resistance loci across Solanaceae and for a large genetic diversity. Theor. Appl. Genet. 106, 1473–1485. [DOI] [PubMed] [Google Scholar]
- Tompkins, C.M. (1937) Phytophthora rot of honeydew melon. J. Agric. Res. 54, 933–944. [Google Scholar]
- Torto, T. , Li, S. , Styer, A. , Huitema, E. , Testa, A. , Gow, N.A.R. , van West, P. and Kamoun, S. (2003) EST mining and functional expression assays identify extracellular effector proteins from Phytophthora . Genome Res. 13, 1675–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit, S. , Worle, J.M. , Nurnberger, T. , Koch, W. and Seitz, H.U. (2001) A novel protein elicitor (PaNie) from Pythium aphanidermatum induces multiple defense responses in carrot, Arabidopsis, and tobacco. Plant Physiol. 127, 832–841. [PMC free article] [PubMed] [Google Scholar]
- Whisson, S.C. , Boevink, P.C. , Moleleki, L. , Avrova, A.O. , Morales, J.G. , Gilroy, E.M. , Armstrong, M.R. , Grouffaud, S. , van West, P. , Chapman, S. , Hein, I. , Toth, I.K. , Pritchard, L. and Birch, P.R. (2007) A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature, 450, 115–118. [DOI] [PubMed] [Google Scholar]
- Wiant, J.S. (1940) A rot of winter queen watermelons caused by Phytophthora capsici . J. Agric. Res. 60, 73–88. [Google Scholar]


