SUMMARY
Members of the Wheat‐Induced Resistance 1 (TaWIR1) gene family are highly induced in response to a wide range of pathogens. Homologues have been identified in barley, but not in Brachypodium, whereas, in rice, only distant WIR1 candidates are known. Phylogenetic analysis placed TaWIR1a and TaWIR1b within a distinct clade of wheat transcripts, whereas TaWIR1c clustered with HvWIR1 genes. Transcripts of all three TaWIR1 genes were strongly induced by a wheat‐adapted isolate of Magnaporthe oryzae. Virus‐induced gene silencing of the TaWIR1 gene family had no effect on the initial penetration of epidermal cells by M. oryzae. However, following the establishment of an infection site, the fungus was able to grow more extensively within the leaf tissue, relative to control leaves, indicating a role for the TaWIR1 gene family in the cell‐to‐cell movement of M. oryzae. In contrast, the silencing of TaWIR1 transcripts had no effect on epidermal cell penetration by a wheat‐adapted isolate of Blumeria graminis, or on the subsequent growth of hyphae. Differential transcription of TaWIR1 genes was also seen in epidermal peels, relative to the remaining leaf tissue, following inoculation with M. oryzae.
INTRODUCTION
In a world economy faced with global food insecurity, the demand for increased agricultural production has never been greater. Together with rice and maize, wheat provides a substantial proportion of the calorific intake of the human population, either directly or through livestock feed (http://faostat.fao.org). Disease presents a major constraint to wheat production, causing significant yield losses (Kosina et al., 2007). The most effective means of disease control is by the informed use of resistance genes through conventional breeding. A greater understanding of how individual genes contribute to resistance would advise breeders as to which combinations of resistance genes would prove to be most effective, providing long‐term resistance.
Plants have evolved elegant molecular defence mechanisms that can be broadly considered to operate at two stages. The first is an early, broad defence response that recognises potential pathogens through the detection of conserved molecules, termed microbe‐ or pathogen‐associated molecular patterns (MAMPs/PAMPs) (Zipfel, 2009). This receptor‐mediated recognition leads to PAMP‐triggered immunity (PTI), which functions to limit the extent of initial pathogen penetration and invasion (Jones and Dangl, 2006). The second layer of defence involves plant resistance (R) proteins that have evolved to recognise specific pathogen‐derived effector molecules that function to suppress PTI and promote pathogen colonisation. This effector‐triggered immunity (ETI) is typically race specific and is often mediated by nucleotide‐binding site leucine‐rich repeat (NBS‐LRR)‐type R genes (Jones and Dangl, 2006). When a pathogen successfully evades both PTI and ETI, a compatible interaction is established.
In addition to the many race‐specific R genes identified in wheat, resistance loci that confer race‐nonspecific resistance have also been identified (Boyd, 2006; Singh et al., 2011). These genes often confer a partial resistance, and the recent cloning of the well‐characterised leaf rust (causal agent Puccinia triticina) race‐nonspecific resistance Lr34 has identified a gene that encodes for a multidrug ABC transporter (Krattinger et al., 2009). Similarly, in barley, the Rphq11 gene for partial, race‐nonspecific resistance to leaf rust has been cloned, and encodes for a phospholipid hydroperoxide glutathione peroxidase (Chen et al., 2010). The mapping of defence‐related genes in barley, including barley homologues of the Wheat‐Induced Resistance 1 (TaWIR1) genes, has shown co‐localisation with partial resistance loci for leaf rust and powdery mildew, further supporting the value of these defence‐related genes as candidates for partial resistance amenable to resistance breeding (Aghnoum et al., 2010; Douchkov et al., 2011; Marcel et al., 2007).
The TaWIR1 gene family was first identified following inoculation of the wheat cv. Fidel with a nonadapted, barley isolate of the powdery mildew pathogen Blumeria graminis (Schweizer et al., 1989). Two sequences were discovered, TaWIR1a and TaWIR1b (Bull et al., 1992). Subsequently, a third sequence, TaWIR1c, was identified (Franck and Dudler, 1995). TaWIR1 encodes for a small glycine‐ and proline‐rich protein, the N‐terminus of which has a potential signal peptide or membrane‐spanning region. The negative charge difference between the amino acids in the C‐terminal region and those flanking the N‐terminal region suggests that the C‐terminal region is extracytoplasmic. This, together with the abundance of glycine and proline residues, which may support cell wall contact, led Bull et al. (1992) to suggest that these proteins may play a role in enhancing the adhesion of the plasma membrane to the cell wall during pathogen attack.
In wheat, WIR1 transcripts have been shown to steadily increase over time in response to a number of adapted and nonadapted isolates of fungal pathogens, including isolates of Magnaporthe spp. (Tufan et al., 2009), Blumeria spp. (Schweizer et al., 1989), Puccinia striiformis (Bozkurt et al., 2010; 2008a, 2008b, 2010), P. triticina (Bolton et al., 2008) and Fusarium spp. (Desmond et al., 2008; Diethelm et al., 2011; Jia et al., 2009). Homologues of TaWIR1 have only been identified in barley (HvWIR1; Douchkov et al., 2011) and rice (OsRIR1; Mauch et al., 1998). In barley, HvWIR1 transcripts were induced in response to adapted and nonadapted isolates of Blumeria spp. (Jansen et al., 2005; Zierold et al., 2005), and a wheat‐adapted isolate of leaf rust P. triticina (Neu et al., 2003). In rice, the OsRIR1 protein shows only approximately 30% amino acid identity to the predicted TaWIR1 protein (Mauch et al., 1998; Yuan et al., 2004). OsRIR1 transcripts accumulate on inoculation with adapted M. oryzae isolates, the nonadapted bacterium Pseudomonas syringae pv. syringae and following treatment with the Pseudomonas syringae pv. syringae defence activator peptide syringolin A (Wäspi et al., 1998). TaWIR1‐ and OsRIR1‐like genes have also been reported to respond to feeding by Hessian fly (Mayetiola destructor) larvae in wheat (Sardesai et al., 2005) and brown plant hopper (Nilaparvata lugens) in rice (Yuan et al., 2004), whereas HvWIR1 transcripts have been found in barley phloem sap following bird‐cherry oat aphid (Rhopalosiphum padi) feeding (Gaupels et al., 2008). Unlike TaWIR1, OsRIR1 transcripts are also induced on wounding (Bull et al., 1992; Mauch et al., 1998; Schweizer et al., 1998; Wäspi et al., 1998). This induction of transcription in response to such a broad range of pests and pathogens suggests that TaWIR1, HvWIR1 and OsRIR1 genes are involved in basal resistance in cereals.
Blumeria graminis f. sp. tritici, the causal agent of powdery mildew, is a common field pathogen of wheat (Cunfer, 2002). More recently, wheat blast, causal agent M. oryzae, has occurred as a field disease of wheat in South America, appearing for the first time in Brazil in 1986 (1993, 2004). Both B. graminis f. sp. tritici and M. oryzae form appressoria on the leaf surface and enter an epidermal cell via an infection peg which breaches the plant cell wall (Boyd et al., 1995; Tufan et al., 2009). Blumeria graminis does not invade past the epidermis, haustoria being the only fungal structures formed within plant cells, with all subsequent hyphal growth occurring on the leaf surface (Boyd et al., 1995). Magnaporthe oryzae hyphae invade both epidermal and mesophyll tissue, spreading from the initial infected epidermal cell into neighbouring epidermal and mesophyll cells through plasmodesmata (Kankanala et al., 2007). An in‐depth histopathological study of the interaction between wheat and M. oryzae has been reported by Tufan et al. (2009).
In this study, we investigate the genomic complexity of the TaWIR1 gene family in the hexaploid wheat cv. Renan, and the relationship of members of the TaWIR1 gene family to WIR1 homologues in other plant species. The role of the different members of the TaWIR1 gene family in restricting fungal pathogen infection in wheat is addressed through transient gene silencing using the Barley stripe mosaic virus (BSMV)‐mediated virus‐induced gene silencing (VIGS) system and microscopy to follow pathogen development. The fungal pathogens causing wheat blast, M. oryzae, and powdery mildew, B. graminis f. sp. tritici, are examined to compare the influence of TaWIR1 gene expression on the initial penetration of an epidermal cell by the fungus, a stage common to both M. oryzae and B. graminis f. sp. tritici, and on the cell‐to‐cell movement of the fungal pathogen, for M. oryzae only.
RESULTS
Expression of TaWIR1 transcripts in response to fungal pathogen inoculation
Transcript analysis of wheat inoculated with Magnaporthe spp. indicated that transcripts of TaWIR1 genes were induced significantly by both adapted and nonadapted isolates of Magnaporthe spp. (Tufan et al., 2009). Published wheat Affymetrix GeneChip microarray data generated from pathogen‐inoculated wheat tissue were used to investigate the expression pattern of TaWIR1 transcripts in other wheat–pathogen interactions. Analysis included species of the fungal pathogens Magnaporthe and Blumeria, P. striiformis and P. triticina, and Fusarium pseudograminearum. Nine probe sets with significant similarity to the TaWIR1a, TaWIR1b and TaWIR1c coding sequences, based on low E‐value scores (<E‐30), were identified on the Affymetrix GeneChip (Table S1, see Supporting Information). Meta‐analysis indicated that, in most cases, TaWIR1 transcripts were up‐regulated from 24 to 48 h after pathogen inoculation in compatible, incompatible and nonhost interactions, reflecting a peak of TaWIR1 gene transcription during the initial stages of attempted pathogen colonisation (Fig. 1).
Figure 1.
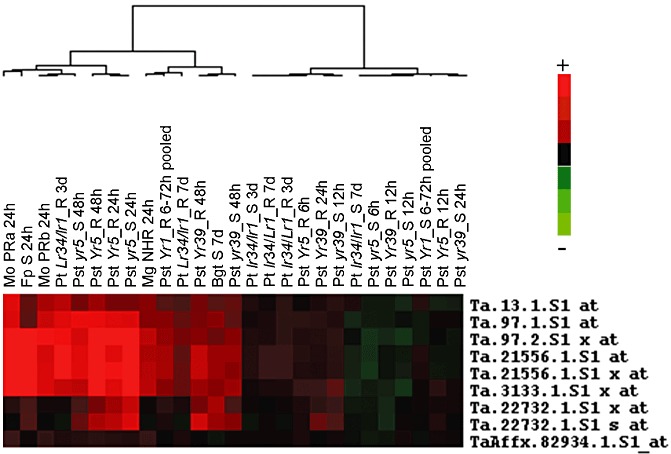
Meta‐analysis of TaWIR1 probe sets on the Affymetrix Wheat GeneChip differentially transcribed in response to fungal pathogen inoculation. Experiment descriptions are shown at the top of the figure, with the probe set names listed on the right of the figure. Experiments included are Magnaporthe oryzae (Mo) inoculations with isolate BR32 (a) or BR37 (b), partial resistance (PR) interactions, with M. grisea (Mg) isolate BR29, a nonhost interaction (NHR) (Tufan et al., 2009), incompatible and compatible interactions with Puccinia striiformis f. sp. tritici (Pst) (Yr5, Yr39, Yr1; 2008a, 2008b and Bozkurt et al., 2010, respectively), incompatible and compatible interactions with P. triticina (Pt) (Lr34 and Lr1; Bolton et al., 2008), and compatible Blumeria graminis f. sp. tritici (Bgt; Chain et al., 2009) and Fusarium pseudograminearum (Fp; Desmond et al., 2008) inoculations. The time points sampled post‐inoculation are shown alongside the pathogen labels. Red blocks indicate transcript up‐regulation and green blocks indicate transcript down‐regulation in pathogen‐inoculated tissue relative to controls. Samples represent leaf‐inoculated tissues, except Fp, which is from inoculated stem bases.
Genetic complexity of the TaWIR1 gene family in wheat
Transcript cloning, sequencing and single‐strand conformation polymorphism (SSCP) analysis were used to investigate the genetic complexity of the WIR1 gene family in wheat cv. Renan, together with phylogenetic analyses of all WIR1‐like sequences retrieved in silico. Polymerase chain reaction (PCR) primers designed specifically to the coding sequences of TaWIR1a (GenBank accession number M95500), TaWIR1b (M94959) and TaWIR1c (X87686; Table S2, see Supporting Information) were used to amplify full‐length cDNAs from cv. Renan post‐inoculation with an isolate of M. oryzae to which Renan shows partial resistance. SSCP analysis of these cDNAs indicated that TaWIR1a and TaWIR1b each produced a single transcript in Renan, whereas at least two transcripts were amplified using the primers for TaWIR1c (Fig. 2A).
Figure 2.
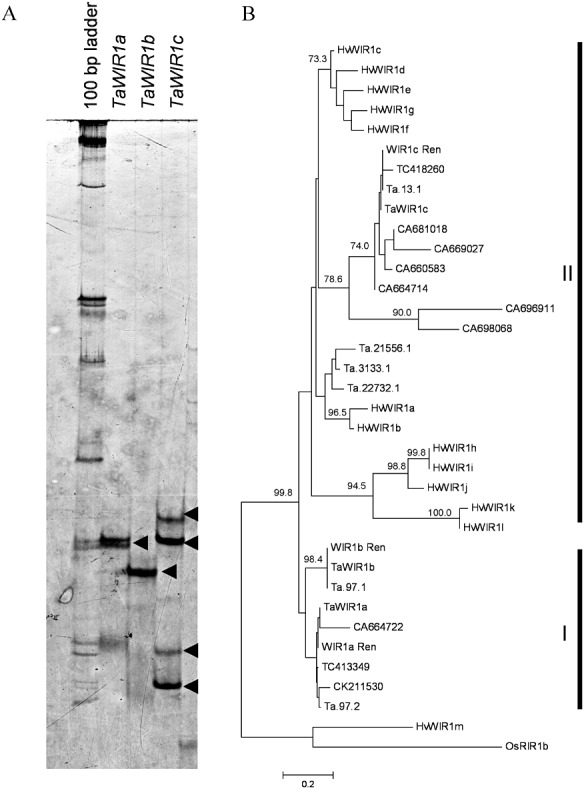
Molecular analysis of the TaWIR1 gene family in wheat. (A) Single‐strand conformation polymorphism (SSCP) analysis of TaWIR1a, TaWIR1b and TaWIR1c transcripts amplified from the wheat cv. Renan. (B) Phylogenetic analysis of TaWIR1 and HvWIR1 proteins using the neighbour‐joining method in phylip. Confidence levels are given above the nodes, obtained from a 1000 bootstrap analysis. The rice WIR1‐like protein, OsRIR1b, was used as an outlier. The sequences shown represent the wheat Affymetrix probe sets (Ta), the Dana Farber Cancer Institute (DFCI) tentative consensus (TC) sequences and GenBank accessions. The GenBank accession numbers for the published TaWIR1 and OsRIR1b sequences are: TaWIR1a (M95500), TaWIR1b (M94959), TaWIR1c (X87686) and OsRIR1b (Y14824).
Comparative analysis of the DNA sequences of clones of TaWIR1a, TaWIR1b and TaWIR1c indicated that TaWIR1a and TaWIR1b transcripts from Renan were identical to the published nucleotide sequences from wheat cv. Fidel. Although SSCP analysis identified two possible transcript sequences, only a single TaWIR1c sequence was cloned from Renan. The reason for this was not clear, but may indicate alternative transcript splicing that was not detected by cloning and sequencing. The sequence differed from the published TaWIR1c sequence by a three‐nucleotide insertion (CGG after position 116) and a single nucleotide substitution (T→C position 186). The three‐nucleotide insertion resulted in a single amino acid insertion in Renan (G after position 35), whereas the nucleotide substitution at position 186 was silent. The full‐length coding sequences of TaWIR1a, TaWIR1b and TaWIR1c from wheat cv. Renan have been deposited in the GenBank database under the accession numbers HQ337015, HQ337016 and HQ337017.
Similarity searches using the DNA sequences of the three published TaWIR1 genes identified up to 15 TaWIR1‐like transcripts in wheat (Table S3, see Supporting Information). A number of barley homologues (Douchkov et al., 2011) and distant rice WIR1 candidates (Mauch et al., 1998; Schaffrath et al., 2000) have been described; however, no WIR1‐like transcripts were identified in the Brachypodium genome using either the blastn or tblastx search tools (http://blast.brachypodium.org/). Phylogenetic analysis, using the predicted amino acid sequences of the WIR1‐like transcripts found in wheat and barley, indicated that TaWIR1a and TaWIR1b were distinct from the barley HvWIR1 transcripts (Douchkov et al., 2011; Fig. 2B). The WIR1‐like transcripts were initially divided into two clades. The first clade contained the TaWIR1a‐like and TaWIR1b‐like proteins, which were further subdivided into two separate groups. The second clade contained all the other TaWIR1 proteins, including TaWIR1c and all of the HvWIR1 proteins, except HvWIR1m, which formed an outgroup together with the rice protein RIR1b. Using SignalP, all TaWIR1 proteins were predicted to contain a signal peptide, although the probability of cleavage was lower for the TaWIR1a group of proteins (Table S4, see Supporting Information). Subcellular localisation predictions using TargetP indicated that TaWIR1b (clade I) and almost all of the TaWIR1 proteins in clade II, including TaWIR1c, were predicted to be secreted (Table S5, see Supporting Information). Transmembrane helix prediction (TMHMM) analysis predicted a single transmembrane domain for all of the TaWIR1 proteins analysed (Table S6, see Supporting Information). However, the probability of N‐terminus localisation on the cytoplasmic side of the membrane was low (P < 0.15) for all of the TaWIR1a‐like proteins (TaWIR1a, Ta.97.2, CK211530, CA664722, TC413349), suggesting that the C‐termini of these proteins are likely to be found on the cytoplasmic side of the membrane. Together, the low probability scores for cleavage site and cytoplasmic N‐terminus localisation for the TaWIR1a proteins support the likelihood that these proteins may be retained in a membrane.
Effect of BSMV‐mediated VIGS of TaWIR1 on resistance to M. oryzae and B. graminis f. sp. tritici
To silence transcripts of the TaWIR1 gene family, a 159‐bp fragment was designed to a region of the TaWIR1b coding sequence which was relatively conserved in all of the TaWIR1 sequences identified in this study (Fig. S1, see Supporting Information). The silencing fragment exhibited approximately 80% identity to all of the identified TaWIR1 sequences, except Ta.21556.1, Ta.3133.1, Ta.22732.1, CA669027 and CA696911 (Table S7, see Supporting Information). The 159‐bp TaWIR1 VIGS fragment was cloned in antisense orientation, 3′ of the γb gene (BSMV:WIR1; Holzberg et al., 2002). The silencing target prediction software si‐Fi v1.4.0 (http://labtools.ipk‐gatersleben.de/) was used to determine which wheat sequences would be potential targets for silencing by the BSMV:WIR1 construct. Wheat sequences with similarity to TaWIR1a and TaWIR1b (Fig. 2, clade I) were predicted to be efficiently silenced by the BSMV:WIR1 construct (Table S8, see Supporting Information). The only transcript from clade II predicted to be potentially silenced was CA698068. No other wheat sequences were predicted to be silenced by the BSMV:WIR1 construct, indicating that the 159‐bp VIGS fragment was specific to the TaWIR1 gene family.
The first and second leaves of 10‐day‐old Renan seedlings were rub inoculated with BSMV:WIR1 or BSMV:GFP (GFP, green fluorescent protein), or mock inoculated with FES buffer (sodium‐pyrophospate 1% w/v; macaloid 1% w/v; celite 1% w/v; 0.5 m glycine; 0.3 m K2HPO4, pH 8.5). TaWIR1 transcript levels were then monitored, 14 days post‐inoculation (dpi), by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) using primer sets specific for TaWIR1a, TaWIR1b and TaWIR1c. The transcript levels for all three TaWIR1 genes were reduced in BSMV:WIR1‐inoculated plants (Fig. 3). Although the BSMV:GFP VIGS control was shown to induce TaWIR1 genes, in seedlings inoculated with BSMV:WIR1, the VIGS mechanism still effectively reduced transcript levels to well below the levels seen in mock‐inoculated leaf tissue. Furthermore, transcripts of Ta.21556.1, Ta.3133.1 and Ta.22732.1, which showed less than 80% nucleotide identity to the BSMV:WIR1 VIGS insert, also exhibited suppressed transcript levels in BSMV:WIR1‐inoculated plants (Fig. S2, see Supporting Information).
Figure 3.
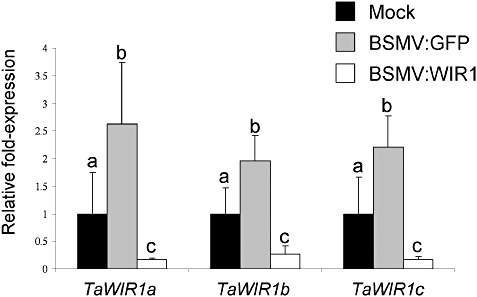
Transcript levels of the TaWIR1 genes in Barley stripe mosaic virus (BSMV)‐mediated virus‐induced gene silenced wheat seedlings of cv. Renan. Mean transcript levels of TaWIR1a, TaWIR1b and TaWIR1c, relative to mock‐inoculated controls, are shown for two independent experiments, consisting of three biological replicates, inoculated with BSMV:WIR1 or the BSMV:GFP control. Relative transcript levels which do not differ significantly from each other are represented by the same lower case letter. Error bars show standard errors.
The effects of silencing of the TaWIR1 gene family on resistance in wheat cv. Renan was analysed for the fungal pathogens M. oryzae and B. graminis f. sp. tritici. For both pathogens, analysis of the infection process was carried out in BSMV:WIR1‐inoculated seedlings in which TaWIR1 transcript levels were confirmed to be reduced by qRT‐PCR in the adjacent leaf. M. oryzae isolate BR32 was spray inoculated onto Renan seedlings at 14 dpi with the VIGS construct BSMV:WIR1 or the viral construct control BSMV:GFP. The interaction between Renan and isolate BR32 was assessed 96 h after fungal inoculation by comparing the proportion of infection sites at three different stages of the wheat–Magnaporthe interaction: HALO, HYPFLUO and MULTIFLUO (see Experimental procedures) (Fig. 4; Tufan et al., 2009). In BSMV:WIR1‐inoculated plants, there were significantly more infection sites at which M. oryzae hyphae had invaded multiple cells (MULTIFLUO; t‐test probability, 0.038) and significantly fewer infection sites at which hyphae were restricted to the first invaded epidermal cell (HYPFLUO; t‐test probability, 0.057), relative to BSMV:GFP‐inoculated plants. No significant differences (t‐test probability, 0.362) were found in the number of infection sites halted at the HALO stage (Fig. 4A). This suggests that TaWIR1 does not play a role in pre‐penetration resistance to M. oryzae, but that silencing of the TaWIR1 genes enabled increased intracellular spread of isolate BR32.
Figure 4.
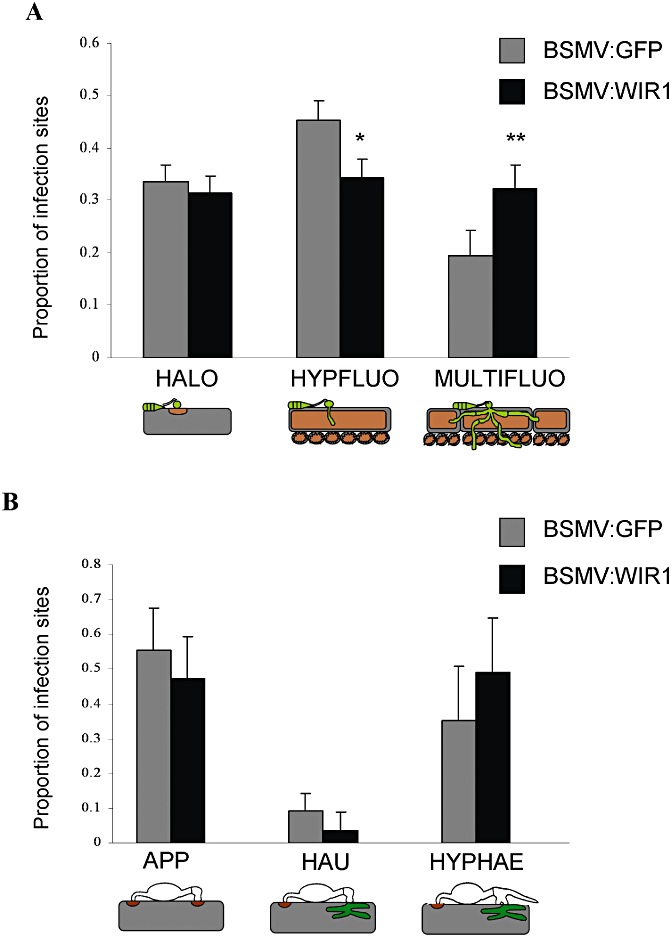
Proportion of fungal growth stages on Barley stripe mosaic virus (BSMV)‐mediated virus‐induced gene silenced seedlings of the wheat cv. Renan at 96 h post‐inoculation (hpi) with Magnaporthe oryzae (A) and 48 hpi with Blumeria graminis f. sp. tritici (B). Each growth stage is shown relative to the number of infection sites. For M. oryzae, growth stages were defined as sites that had failed to establish hyphae, attempted penetration being associated with the accumulation of autofluorescent compounds beneath the appressorium (HALO), as sites at which hyphae had invaded the first epidermal cell, being associated with plant cell autofluorescence (HYPFLUO), and as sites of multiple cell invasion associated with autofluorescence (MULTIFLUO). For B. graminis f. sp. tritici, growth stages were defined as appressoria that had failed to establish a haustorium (APP), as haustoria within epidermal cells, but lacking surface hyphae (HAU), and where hyphae had formed on the leaf surface (HYPHAE). Mean values are shown from three independent experiments. Significant differences between growth stages are shown at t‐test probabilities of >0.01 (**) and >0.1 (*). Error bars show standard errors.
The B. graminis f. sp. tritici isolate JIW2 was inoculated onto Renan seedlings at 14 dpi with the VIGS construct BSMV:WIR1 or the viral control BSMV:GFP. The effects of silencing of the TaWIR1 genes on isolate JIW2 development was assessed 48 h after fungal inoculation by measuring the proportion of infection sites at three different stages of the wheat–powdery mildew interaction: APP, HAU and HYPHAE (see Experimental procedures) (Fig. 4B; Tufan et al., 2011). There were no significant differences between BSMV:WIR1‐silenced plants and plants inoculated with BSMV:GFP with regard to the proportion of infection sites at each of the three stages: APP (t‐test probability, 0.643), HAU (t‐test probability, 0.485) or HYPHAE (t‐test probability, 0.555). In addition, no significant differences were found in the number of powdery mildew colonies observed on BSMV:WIR1‐ (t‐test probability, 0.779) or BSMV:GFP‐ (t‐test probability, 0.661) inoculated Renan seedlings at 10 dpi with B. graminis f. sp. tritici isolate JIW2, relative to mock‐inoculated control leaves (data not shown). This suggests that silencing of the TaWIR1 genes has no effect on the early development of B. graminis f. sp. tritici on wheat cv. Renan, or on subsequent colony development.
Spatial expression of TaWIR1 transcripts in wheat leaf tissue following inoculation with M. oryzae
TaWIR1 transcript levels were measured in epidermal peels, in the remaining leaf tissue minus the abaxial epidermis (referred to as the mesophyll region) and in intact leaves at 48 h post‐inoculation (hpi) with M. oryzae isolate BR32 or 0.25% gelatine solution, the mock‐inoculated control. The 48 h time point was chosen as previous studies have shown TaWIR1 transcripts to be highly induced at this time point in intact leaves inoculated with the M. oryzae isolate BR32 (Fig. S3, see Supporting Information; Tufan et al., 2009). Following inoculation with BR32, all three TaWIR1 genes were induced in both regions of the leaf, relative to the same region in the mock‐inoculated control (Fig. S4, see Supporting Information). Comparison of the transcript levels of TaWIR1a, TaWIR1b and TaWIR1c in epidermal peels and in the mesophyll region, relative to the transcript levels in the intact leaf, in three independent experiments, provided a measure of the relative expression of each gene in these two regions of the leaf (Table 1). Transcript levels of TaWIR1a and TaWIR1c in BR32‐inoculated leaves were higher in the mesophyll region of the leaf relative to the abaxial epidermal tissue at 48 hpi, whereas transcript levels of TaWIR1b were not significantly different between the two regions of the leaf. In the mock‐inoculated control, transcript levels of all three genes were similar across the leaf (Table 1). TaRUBISCO (RUBISCO, ribulose‐1,5‐bisphosphate carboxylase oxygenase) expression was not induced by M. oryzae inoculation, and confirmed the quality of the epidermal peel tissue, with significantly less TaRUBISCO transcripts being found in the epidermis relative to the remaining leaf tissue (Table 1).
Table 1.
Gene transcript levels in leaf tissues inoculated with Magnaporthe oryzae isolate BR32 or mock inoculated with FES buffer, relative to the transcript levels in the intact leaf.
| Tissue | TaWIR1a ‡ | TaWIR1b | TaWIR1c | TaRUBISCO |
|---|---|---|---|---|
| Mock | ||||
| Epidermis* | 0.69 a | 1.06 a | 0.89 a , b | 0.28 b |
| (0.44) | (0.47) | (0.24) | (0.09) | |
| Mesophyll† | 1.13 a | 0.80 a | 1.35 a | 1.12 a |
| (0.46) | (0.45) | (0.57) | (0.23) | |
| BR32 | ||||
| Epidermis | 0.28 b | 0.59 a | 0.32 c | 0.24 b |
| (0.15) | (0.26) | (0.16) | (0.03) | |
| Mesophyll | 0.80 a | 0.82 a | 0.86 b | 1.29 a |
| (0.58) | (0.34) | (0.59) | (0.18) |
Epidermal peels of the abaxial leaf layer.
The remaining leaf tissue minus the abaxial epidermal layer.
The mean relative transcript levels from three replicate experiments are shown for each gene. For each gene, mean values showing the same letter do not differ significantly at a t‐test probability of <0.001.
Standard deviations of the means are shown in parentheses.
DISCUSSION
WIR1 was first described in wheat, being induced by the fungal pathogen responsible for powdery mildew, B. graminis (Schweizer et al., 1989). Homologues of TaWIR1 have been found in barley, as well as a distant candidate in rice, OsRIR1. No candidate for WIR1 was found in the Brachypodium genome sequence. WIR1 transcripts are induced in wheat and barley following inoculation with a diverse range of pathogenic organisms (Bolton et al., 2008; Bozkurt et al., 2010; Bull et al., 1992; 2008a, 2008b, 2010; Desmond et al., 2008; Jia et al., 2009; Schweizer et al., 1989; Tufan et al., 2009), but not by abiotic stress (Douchkov et al., 2011). In barley, HvWIR1 transcripts have also been found to accumulate in noninoculated leaves of adult plants (Douchkov et al., 2011), which may indicate a link with the developmentally regulated, broad‐spectrum, partial resistance often seen as plants mature (Boyd, 2006). Four HvWIR1 loci have been shown to co‐locate with quantitative trait loci conferring partial resistance to B. graminis f. sp. hordei in barley (Aghnoum et al., 2010; Douchkov et al., 2011). This association was further supported using single nucleotide polymorphisms (SNPs) within HvWIR1 genes in a genetic association analysis of resistance towards B. graminis f. sp. hordei (Comadran et al., 2009; Douchkov et al., 2011). In rice, overexpression of the TaWIR1 homologue, OsRIR1b, has also been shown to enhance resistance to M. oryzae (Schaffrath et al., 2000). The WIR1 genes are therefore good candidates for a resistance mechanism which supports broad‐spectrum resistance. However, the mechanism by which the WIR1 proteins deliver this resistance remains to be determined.
Although similarity searches identified up to 15 TaWIR1‐like transcripts, phylogenetic analysis grouped the TaWIR1a and TaWIR1b gene sequences into a distinct clade, whereas the TaWIR1c sequence clustered together with the barley HvWIR1 homologues. A Southern analysis in wheat cv. Fidel identified up to six bands detected by the TaWIR1 sequence (Bull et al., 1992), whereas cloning of the TaWIR1a, TaWIR1b and TaWIR1c transcripts from cv. Renan only identified three sequences. The TaWIR1a and TaWIR1b gene sequences from Renan were also identical to the published sequences cloned from cv. Fidel. Mapping of the TaWIR1a and TaWIR1b sequences in wheat identified a single locus for both genes, located on chromosomes 7BS and 5DS, respectively (Diethelm et al., 2011). Collectively, these data suggest a relatively simple genomic structure for the TaWIR1 genes, with a high degree of sequence conservation. The diversity of TaWIR1‐like sequences found in database searches probably reflects gene sequence variation between different wheat varieties. Subsequent analysis of the wheat 454 genomic sequence data, variety Chinese Spring (http://www.cerealsdb.uk.net/CerealsDB/Documents/DOC_CerealsDB.php), supported our observations, indicating that a single locus of each TaWIR1 gene probably exists on each of the three homoeologous genomes, whereas, between Chinese Spring and cv. Fidel, two to three potential varietal SNPs were identified within the TaWIR1 genes.
Silencing of the TaWIR1 genes had no effect on the initial penetration of epidemal cells by the adapted M. oryzae isolate BR32, but facilitated the subsequent intracellular spread of the fungus, resulting in a greater number of infection sites colonizing multiple mesophyll cells by 96 hpi. The predicted protein structure of TaWIR1 indicates a hydrophobic N‐terminus which, in the case of TaWIR1a, has a low probability of cleavage and a greater probability of being membrane bound (Tables S4 and S6). This opens up the possibility that the TaWIR1a protein is membrane anchored, with the hydrophilic C‐terminus being exposed to the acidic apoplast. In the cowpea rust system, expression of early, cell wall‐related defence responses was associated with the maintenance of adhesion between the plant cell wall and the plasma membrane (Mellersh and Heath, 2001). TaWIR1a may therefore be required to maintain cell wall–plasma membrane contact during wheat–M. oryzae infection. As M. oryzae is believed to pass from cell to cell via plasmodesmata (Kankanala et al., 2007), TaWIR1a may play a role in maintaining the cell wall membrane integrity of these structures. The TaWIR1b and TaWIR1c proteins are predicted to contain signal peptides and are probably secreted. Analysis of OsRIR1a in rice protoplasts indicated that this protein was secreted and was probably ionically bound to cell walls (Mauch et al., 1998). It is therefore possible that TaWIR1b and TaWIR1c proteins are also involved in cell wall strengthening, enhancing the physical barrier that helps restrict pathogen colonization. However, further experimentation is required to determine definitively the functions of the TaWIR1 proteins in plant defence.
In contrast, BSMV‐mediated VIGS of TaWIR1 transcripts had no effect on B. graminis f. sp. tritici colonization, despite inoculation with this organism resulting in increased levels of TaWIR1 transcripts (Fig. 1; Bull et al., 1992; Schweizer et al., 1989). In barley, transient induced gene silencing of HvWIR1a reduced the number of B. graminis f. sp. hordei haustoria seen in barley epidermal cells, whereas silencing of the HvWIR1h gene increased haustoria establishment; silencing of HvWIR1m had no effect on haustorial establishment (Douchkov et al., 2011). However, in wheat, transient overexpression of TaWIR1a had no effect on haustoria formation in the wheat–B. graminis f. sp. tritici interaction (Schweizer et al., 1999). This may indicate that the effects of WIR1 on B. graminis are gene specific, and that the global silencing of the TaWIR1 transcripts in Renan cancelled out any WIR1 gene‐specific effects on B. graminis development.
The initial BSMV:WIR1 construct was designed to the TaWIR1b transcript sequence. The si‐Fi software predicted that this construct would silence the TaWIR1b and TaWIR1a sequences from phylogenetic clade I (Fig. 2A), as well as a single transcript (CA698068) from clade II (Table S8). However, qRT‐PCR analysis indicated that TaWIR1 transcripts within clade II, with less than the previously suggested 80% sequence similarity required for successful VIGS (Holzberg et al., 2002), were also suppressed (Table S7; Fig. S2). The reduced level of sequence similarity required for VIGS, together with the optimal size of 120–500 bp for VIGS fragments (Bruun‐Rasmussen et al., 2007; Holzberg et al., 2002), meant that we were unable to effectively silence specific TaWIR1 transcripts.
Following M. oryzae inoculation, different patterns of spatial expression were observed across the leaf for TaWIR1a/c and TaWIR1b. Similarly, in barley, most HvWIR1 transcripts analysed accumulated to greater levels in the intact leaf relative to epidermal tissue; the exception being HvWIR1a, which accumulated almost as strongly in pathogen‐attacked epidermal tissue as in the intact leaf (Douchkov et al., 2011; Zierold et al., 2005). The differences in spatial expression of the TaWIR1 transcripts may therefore facilitate the role of this gene family in defence against pathogens with different lifestyles (biotrophic, hemibiotrophic or necrotrophic) and inhabiting different plant tissues (Fig. 1; Bolton et al., 2008; Bozkurt et al., 2010; Bull et al., 1992; 2008a, 2008b, 2010; Desmond et al., 2008; Schweizer et al., 1989; Tufan et al., 2009).
Although we can only speculate on the function of the TaWIR1 proteins in pathogen defence, accumulating evidence indicates a significant role for this family of genes in cereal disease resistance. The conservation of the WIR1 gene family in wheat and barley indicates a preserved function very specific to these cereal species. However, when comparing wheat with barley, the role of individual WIR1 genes may have diverged, with different members of the gene family specifically targeting different pathogens.
EXPERIMENTAL PROCEDURES
Plant material
Seeds of the winter wheat cv. Renan (Triticum aestivum L.) were provided by Jean‐Benoit Morel (INRA, Montpellier, France). Plants were grown in a peat and sand (1:1) mix at 23 °C in a Fitotron growth cabinet (Sanyo Gallenkamp PLC, Loughborough, UK) with a 16‐h/8‐h light/dark cycle.
Meta‐analysis of TaWIR1 transcripts
The predicted amino acid sequences of the TaWIR1a (Q01482, Bull et al., 1992), TaWIR1b (Q01481, Bull et al., 1992) and TaWIR1c (Q41581, Franck and Dudler, 1995) proteins were retrieved from the National Center for Biotechnology Information (NCBI). Probe sets representing TaWIR1 genes on the Affymetrix Wheat GeneChip were retrieved using the coding sequences of the three TaWIR1 proteins and blastn to query the microarray (Wise et al., 2007). Probe sets with high similarity scores (<E‐30) and defined as not potentially unreliable (Schreiber et al., 2009) were selected for meta‐analysis (Table S1). Wheat Affymetrix datasets for experiments investigating wheat–pathogen interactions were downloaded from PlexDB: TA9 (Coram et al., 2008b), TA11 (Coram et al., 2008a), TA24 (Tufan et al., 2009), TA25 (Bozkurt et al., 2010), TA31 (Desmond et al., 2008), TA32 (Bolton et al., 2008) and TA34 (Chain et al., 2009). Data were analysed in R using the package AffylmGUI (Wettenhall et al., 2006) following Robust Multichip Average (RMA) normalization (Irizarry et al., 2003). Differential expression was calculated using linear models and an empirical Bayes‐moderated t‐statistic (Smyth, 2004). Contrasts between pathogen‐inoculated and mock‐inoculated control samples were tested. Differential regulation of probe sets was assessed based on expression, the data being exported into a tab‐delimited file. Expression patterns of WIR1 probe sets were analysed using Cluster 3 (Eisen et al., 1998) with a Euclidean distance matrix and complete‐linkage clustering. Data were visualized with Treeview v.1.0.13.
Sequence and phylogenetic analysis of TaWIR1
Primers were designed to the published sequences of TaWIR1a, TaWIR1b and TaWIR1c to amplify the entire coding sequence of each gene from Renan. RNA was extracted from leaves of Renan at 48 hpi with M. oryzae isolate BR32, processed and reverse transcribed as described by Tufan et al. (2009). Transcripts were amplified using Hotstar master mix (Qiagen, Hilden, Germany), following the manufacturer's recommendations. A G‐Storm GS1 thermocycler was used for target amplifcation (GRI, Braintree, Essex, UK), with an initial activation step at 95 °C for 15 min, followed by 35 cycles of 1 min at 94 °C, 1 min at 60 °C for TaWIR1a and TaWIR1b or 68 °C for TaWIR1c, 1 min at 72 °C and a final 10‐min extension cycle. PCR products were purified using a QIAquick PCR purification kit (Qiagen) and cloned into pGEM‐T Easy (Promega, Madison, WI, USA). Plasmid DNA was isolated from more than 10 independent clones for each TaWIR1 gene using a QIAprep spin Miniprep kit (Qiagen) and sequenced by Genome Enterprise Ltd (http://orders2.genome‐enterprise.com/).
Using blastn, the published TaWIR1 coding sequences were used to query the Dana Farber Cancer Institute (DFCI) gene indices database (http://compbio.dfci.harvard.edu/tgi/cgi‐bin/tgi/Blast/index.cgi) to recover similar wheat tentative consensus (TC) sequences, as defined by low E‐value scores (<E‐30). Sequences that had identical coding sequences to the published TaWIR1 sequences, to each other or that showed incomplete open reading frames, i.e. no start codon or multiple stop codons, were excluded. Selected TC sequences and Affymetrix GeneChip probe sets representing TaWIR1 genes were translated and aligned using prankster (Löytynoja and Goldman, 2008) to the published amino acid sequences of TaWIR1a, TaWIR1b and TaWIR1c, together with the predicted protein sequences of these genes from Renan. Amino acid sequences of barley HvWIR1 transcripts (Douchkov et al., 2011) and the rice OsRIR1b gene (Mauch et al., 1998) were included in the alignment.
Phylogenetic analysis was performed using phylip (http://evolution.genetics.washington.edu/phylip.html), and a phylogenetic tree was constructed using the neighbour‐joining method, with OsRIR1b as an outgroup. The tree was visualized in mega (Kumar et al., 2008). Percentage confidence levels were obtained after 1000 bootstraps. Signal peptide and subcellular targeting predictions were made using SignalP and TargetP, respectively (Emanuelsson et al., 2007). Transmembrane predictions were made using TMHMM server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM‐2.0/).
SSCP analysis
Sequence polymorphisms in the TaWIR1a, TaWIR1b and TaWIR1c coding sequences recovered from Renan were examined by SSCP analysis based on the method of Martins‐Lopes et al. (2001) with the following modifications. TaWIR1 coding sequences were amplified by PCR as described above, 4 µL of the product was mixed with loading dye [98% formamide; 10 mm ethylenediaminetetraacetic acid (EDTA); 0.1% (w/v) bromophenol blue; 0.1% (w/v) xylene cyanol] and denatured at 95 °C for 2 min. Following incubation on ice for 1 min, 6 µL of each sample were loaded onto the SSCP gel (390 mm × 300 mm × 0.3 mm) containing 12.5 mL of 2 × MDE (Lonza, Rockland, ME, USA), 1.5 mL of 20 × 1.78M Tris‐base, 0.57 m taurine, 10 mm Na2‐EDTA (TTE), 9 mL of 50% glycerol and 27 mL of water, and polymerized by adding 303 µL of 10% (w/v) ammonium persulphate and 27 µL N, N, N', N'‐tetramethylethylenediamine (TEMED). Samples were electrophoresed in a 0.6 × TTE running buffer for at least 16 h.
Silver staining of DNA was performed by passing the polyacrylamide gels through the following steps: fixation for 30 min in 10% acetic acid; washing in double‐distilled H2O for 10 min; staining in silver nitrate solution [5.7 mm silver nitrate plus 0.15% (v/v) formaldehyde] for 30 min; and placing in developer [0.73 m sodium carbonate, 0.15% (v/v) formaldehyde plus 15 mm sodium thiosulphate], chilled to 4 °C, until bands were clearly visible. The reaction was stopped by the addition of 10% acetic acid to the developer.
BSMV‐mediated VIGS of WIR1 genes in wheat cv. Renan
A 159‐bp fragment corresponding to a relatively conserved region between TaWIR1 gene sequences was amplified from Renan cDNA using gene‐specific primers (Table S2), incorporating NotI and PacI restriction enzyme sites at their terminal ends. The phytoene desaturase (PDS) fragment was removed from the BSMVγ.bPDSas vector by digestion with NotI and PacI, and replaced with the TaWIR1 PCR fragment, generating the construct BSMVγ:WIR1as (BSMV:WIR1; Hein et al., 2005; Holzberg et al., 2002).
Purified plasmids containing the BSMV α, β and γ genomes were linearized using endonucleases MluI for α, SpeI for β, BssHII for γWIR1 and SwaI for γGFP. Viral RNA was synthesized from linearized plasmids using an mMessage mMachine T7 in vitro transcription kit (Ambion, Austin, TX, USA). BSMV inoculum was prepared by adding 1 µL of RNA of each viral genome to 27 µL of FES buffer (Pogue et al., 1998). First and second leaves of 10‐day‐old Renan seedlings were rub inoculated with 30 µL of this mixture (Scofield et al., 2005). Plants were rinsed with water to remove excess inoculum and transferred to a 25 °C growth chamber. Secondary pathogen inoculations were performed at 14 dpi with BSMV. Three independent VIGS experiments, each consisting of three biological replicates, were carried out.
si‐Fi analysis to predict potential off‐target silencing effects
To test for potential off‐target silencing effects of the TaWIR1 VIGS fragment, si‐Fi software v1.4.0 (http://labtools.ipk‐gatersleben.de/) was used. The TaWIR1 VIGS fragment was queried against databases downloaded from HarvEST v. 1.56 (http://harvest.ucr.edu/) and Triticum aestivum transcript assembly release 2 (ftp://ftp.tigr.org/pub/data/plantta/Triticum_aestivum/) using default settings. Any returned sequence that had hits to the TaWIR1 VIGS fragment was considered to be a potential off‐target for gene silencing, although some of the hits were not scored as efficient, suggesting that silencing in these cases may not be complete (Douchkov et al., 2011).
qRT‐PCR analysis
Transcript levels were determined as described by Tufan et al. (2009) using sequence‐specific primers (Table S2). Total RNA was extracted using the RNeasy Plant Mini kit (Qiagen) and genomic DNA contamination was removed with TURBO DNAse I treatment (Ambion). cDNA was prepared from 1 µg of RNA using the Superscript III reverse transcription system (Invitrogen, Carlsbad, CA, USA), primed with random hexamers. Transcripts were amplified with Sybr Green JumpStart™ Taq Ready mix (Sigma, St. Louis, MO, USA) using a DNA engine Opticon2 Continuous Fluorescence Detector (M.J. Research Inc., Alameda, CA, USA), with an initial activation step at 95 °C for 2 min, followed by 40 cycles of 30 s at 95 °C, 30 s at 60 °C and 30 s at 72 °C. Melt curve analysis was performed at the end of each reaction to monitor primer–dimer formation and the amplification of gene‐specific products. Expression levels were normalized using normalization factors derived from geNorm analysis (Vandesompele et al., 2002), employing two or three reference genes (Table S2; Bozkurt et al., 2010; Tufan et al., 2009) that were shown to be stably expressed under the experimental conditions tested. Transcript levels are shown relative to mock‐inoculated control samples, unless stated otherwise.
Magnaporthe oryzae and B. graminis inoculations of BSMV‐VIGS seedlings of cv. Renan
Magnaporthe oryzae isolate BR32 was obtained from Didier Tharreau (CIRAD, Montpellier, France) and was cultured as described previously (Tufan et al., 2009). Wheat powdery mildew (B. graminis f. sp. tritici) isolate JIW2 was obtained from James Brown (John Innes Centre, Norwich, UK). The B. graminis f. sp. tritici isolate was maintained on detached leaf segments of the susceptible wheat cv. Cerco (Boyd et al., 1994).
Magnaporthe oryzae inoculations were carried out as described by Tufan et al. (2009). Wheat seedlings were spray inoculated with 105 spores/mL inoculum. After inoculation, plants were kept at 25 °C and the final symptoms were assessed at 96 hpi. Blumeria graminis f. sp. tritici inoculations were carried out as detached leaf tests (Boyd et al., 1994). Leaf tissue was cut into 3–4‐cm segments and placed, adaxial side up, on water agar (6 g/L) plates supplemented with benzimidazole (0.1 g/L). Spores were blown onto the leaf segments and incubated at 15 °C. Final symptoms were assessed at 10 dpi.
Microscopic and histopathologic analyses of M. oryzae and B. graminis
Clearing, fixing and staining with the fluorescent stain Uvitex‐2B (Ciba‐Geigy, Basle, Switzerland) of M. oryzae‐inoculated leaf tissue were carried out as described by Tufan et al. (2009). Leaf tissue was immersed in chloral hydrate solution (300 mL of 95% ethanol, 125 mL of 90% lactic acid, 800 g of chloral hydrate, made up to 1 L with chloroform), periodically refreshing the solution, for up to 5 days. Leaf samples were stored in lactoglycerol (1:1:1, v/v/v, lactic acid : glycerol : water) at 4 °C. Leaf samples were mounted onto glass slides in 40% glycerol and stored in the dark.
Fungal and plant autofluorescent cellular structures were observed on a Zeiss LSM 510 META confocal microscope (Jena, Germany) using 25× LD LCI Plan‐Apochromat (numerical aperture, 0.8) or 40× EC Plan‐Neofluar (numerical aperture, 1.3) oil immersion objective lenses. Spectral data were collected by excitation with 488‐nm argon (30 mW) and 405‐nm diode (30 mW) lasers. Uvitex‐2B‐stained fungal structures and plant autofluorescing cellular structures were differentiated using a Uvitex‐2B‐specific filter (band pass, 420–480 nm) and an autofluorescence‐specific filter (long pass, 530 nm), respectively.
For microscopic observation of B. graminis f. sp. tritici development, infected leaf segments were cleared and fixed by heating at 70 °C for 1 h in 3:1 ethanol : water (v/v), followed by staining with aniline blue (0.5% w/v in lactoglycerol). Pathogen development was observed using a bright‐field microscope (Nikon 800 Eclipse; Nikon Precision Europe GmbH, Langen, Germany).
Magnaporthe oryzae infection sites were defined by the formation of an appressorium. Fungal growth stages were then defined by the proportion of infection sites: (i) that failed to establish hyphae within the invaded epidermal cell, this attempted penetration usually being associated with the accumulation of autofluorescent compounds beneath the appressorium (HALO); (ii) where hyphae had invaded the first epidermal cell and were associated with epidermal and mesophyll autofluorescence (HYPFLUO); and (iii) that had invaded multiple cells, being associated with epidermal and mesophyll cell autofluorescence in colonised as well as adjacent, noncolonised cells (MULTIFLUO). From each independent silencing experiment, 3–4 cm of leaf tissue, from at least two seedlings, were prepared for analysis. Approximately 100 infection sites were scored in each experiment.
Blumeria graminis f. sp. tritici infection sites were defined by the formation of an appressorium off the appressorium germ tube (Boyd et al., 1995). Fungal growth stages were then defined by the proportion of infection sites: (i) that had failed to establish a haustorium in the underlying epidermal cell, this attempted penetration occasionally being associated with the formation of a papilla beneath the appressorium (APP); (ii) had formed a haustorium within the epidermal cell (HAU); and (iii) had formed hyphae on the leaf surface (HYPHAE) (Boyd et al., 1995). Infection sites associated with long cells were excluded from the analysis. From each independent silencing experiment, 3–4 cm of leaf tissue, from at least two seedlings, were prepared for analysis. Approximately 60 infection sites were scored in each experiment. Blumeria graminis f. sp. tritici colonies were counted at 10 dpi on leaf segments from mock‐inoculated seedlings or seedlings inoculated with BSMV:GFP or BSMV:WIR1, in each of the three independent silencing experiments.
Tissue‐specific expression of wheat TaWIR1 genes
Ten‐day‐old seedlings of cv. Renan were inoculated on both the abaxial and adaxial sides of the leaf with M. oryzae isolate BR32 or mock inoculated with 0.25% gelatine solution. The abaxial epidermis was removed at 48 hpi (Zellerhoff et al., 2010) and total RNA was extracted separately from this epidermal peel tissue, from the remaining leaf tissue and from inoculated whole leaves. Transcript levels of TaWIR1a, TaWIR1b, TaWIR1c and the TaRUBISCO genes were measured by qRT‐PCR using the primers listed in Table S2. TaRUBISCO (accession number AB042066.1) was used to assess mesophyll contamination of the epidermal peels (Eichmann et al., 2010). Three independent epidermal peel experiments, each consisting of at least 100 Renan seedlings per treatment, were carried out.
Statistical analysis
Histopathological analysis of pathogen development was analysed using generalized linear mixed modelling (GLMM; Welham, 1993). A binomial distribution with a logit transformation was used to compare the ratio of the growth stages HALO, HYPFLUO and MULTIFLUO, and APP, HAU and HYPHAE, with the total number of infection sites observed. The model fitted compared replicate experiments and BSMV construct or mock treatments for each pathogen and growth stage. Differences between BSMV treatments significant at an F‐value probability of P < 0.05 were further compared by t‐test analysis.
qRT‐PCR transcript levels were compared using general linear regression. Each TaWIR1 gene was analysed independently. Differences between BSMV VIGS treatments or leaf tissues significant at an F‐value probability of P < 0.05 were further compared by t‐test analysis. All analyses were carried out using genstat for Windows, 12th edition (GenStat Release 12 Committee, 2009).
Supporting information
Fig. S1 CLUSTALW nucleotide alignment of BSMV:WIR1 and TaWIR1 sequences showing conservation of the target region between TaWIR1 genes. Shading indicates 100% (black), 75% (dark grey) and 50% (light grey) conservation between sequences.
Fig. S2 Barley stripe mosaic virus (BSMV)‐mediated virus‐induced gene silencing (VIGS) of TaWIR1 reduces the expression of transcripts corresponding to probe sets Ta.21556.1, Ta.22732.1 and Ta.3133.1 on the Affymetrix Wheat GeneChip, despite these sequence having less than 80% similarity to BSMV:WIR1. Transcript levels were measured by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) and shown relative to mock‐inoculated controls. The mean values of three independent experiments are shown with standard error bars.
Fig. S3 Transcript levels of TaWIR1 genes in intact leaves of cv. Renan measured after inoculation (hours post‐inoculation, hpi) with Magnaporthe oryzae isolate BR32, relative to mock‐inoculated controls. The mean values of two independent experiments are shown with standard error bars.
Fig. S4 Transcript levels of TaWIR1 genes in cv. Renan leaf tissues inoculated with Magnaporthe oryzae isolate BR32, relative to the same region of the leaf in the mock‐inoculated control. The mean values of three independent experiments are shown with standard error bars.
Table S1 TaWIR1 transcripts present as probe sets on the Affymetrix Wheat GeneChip.
Table S2 Primer sequences.
Table S3 TaWIR1 genes identified from database similarity searches.
Table S4 SignalP analysis of TaWIR1 proteins.
Table S5 TargetP analysis of TaWIR1 proteins.
Table S6 Transmembrane prediction for TaWIR1 proteins.
Table S7 Nucleotide identities of TaWIR1 sequences to BSMV:WIR1.
Table S8 BSMV:WIR1 silencing targets predicted by si‐Fi analysis.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
We thank Margaret Corbitt for assistance with powdery mildew inoculations, Ruth Bryant for TaWIR1c primers, Paul Bailey for advice on phylogenetic analysis and Myriam Charpentier for guidance with transmembrane prediction software tools. This work was funded by the Consultative Group on International Agricultural Research, Generation Challenge Project (Cereal Immunity) and by European Research Area network Plant Genomics (TritNONHOST).
REFERENCES
- Aghnoum, R. , Marcel, T.C. , Johrde, A. , Pecchioni, N. , Schweizer, P. and Niks, R.E. (2010) Basal host resistance of barley to powdery mildew: connecting quantitative trait loci and candidate genes. Mol. Plant–Microbe Interact. 23, 91–102. [DOI] [PubMed] [Google Scholar]
- Bolton, M.D. , Kolmer, J.A. , Xu, W.W. and Garvin, D.F. (2008) Lr34‐mediated leaf rust resistance in wheat: transcript profiling reveals a high energetic demand supported by transient recruitment of multiple metabolic pathways. Mol. Plant–Microbe Interact. 21, 1515–1527. [DOI] [PubMed] [Google Scholar]
- Boyd, L.A. (2006) Perspectives: can the durability of resistance be predicted? J. Sci. Food Agric. 86, 2523–2526. [Google Scholar]
- Boyd, L.A. , Smith, P.H. , Green, R.M. and Brown, J.K.M. (1994) The relationship between the expression of defense‐related genes and mildew development in barley. Mol. Plant–Microbe Interact. 7, 401–410. [Google Scholar]
- Boyd, L.A. , Smith, P.H. , Foster, E.M. and Brown, J.K.M. (1995) The effects of allelic variation at the MLA resistance locus in barley on the early development of Erysiphe‐graminis f sp hordei and host responses. Plant J. 7, 959–968. [Google Scholar]
- Bozkurt, T.O. , McGrann, G.R.D. , MacCormack, R. , Boyd, L.A. and Akkaya, M.S. (2010) Cellular and transcriptional responses of wheat during compatible and incompatible race‐specific interactions with Puccinia striiformis f. sp tritici . Mol. Plant Pathol. 11, 625–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruun‐Rasmussen, M. , Madsen, C.T. , Jessing, S. and Albrechtsen, M. (2007) Stability of Barley stripe mosaic virus‐induced gene silencing in barley. Mol. Plant–Microbe Interact. 20, 1323–1331. [DOI] [PubMed] [Google Scholar]
- Bull, J. , Mauch, F. , Hertig, C. , Rebmann, G. and Dudler, R. (1992) Sequence and expression of a wheat gene that encodes a novel protein associated with pathogen defense. Mol. Plant–Microbe Interact. 5, 516–519. [DOI] [PubMed] [Google Scholar]
- Chain, F. , Côté‐Beaulieu, C. , Belzile, F. , Menzies, J.G. and Bélanger, R.R. (2009) A comprehensive transcriptomic analysis of the effect of silicon on wheat plants under control and pathogen stress conditions. Mol. Plant–Microbe Interact. 22, 1323–1330. [DOI] [PubMed] [Google Scholar]
- Chen, X. , Hackett, C.A. , Niks, R.E. , Hedley, P.E. , Booth, C. , Druka, A. , Marcel, T.C. , Vels, A. , Bayer, M. , Milne, I. , Morris, J. , Ramsay, L. , Marshall, D. , Cardle, L. and Waugh, R. (2010) An eQTL analysis of partial resistance to Puccinia hordei in barley. PLoS ONE, 5, e8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comadran, J. , Thomas, W.T.B. , van Eeuwijk, F.A. , Ceccarelli, S. , Grando, S. , Stanca, A.M. , Pecchioni, N. , Akar, T. , Al‐Yassin, A. , Benbelkacem, A. , Ouabbou, H. , Bort, J. , Romagosa, I. , Hackett, C.A. and Russell, J.R.C. (2009) Patterns of genetic diversity and linkage disequilibrium in a highly structured Horedum vulgare association‐mapping population of the Mediterranean basin. Theor. Appl. Genet. 119, 175–187. [DOI] [PubMed] [Google Scholar]
- Coram, T.E. , Settles, M.L. and Chen, X.M. (2008a) Transcriptome analysis of high‐temperature adult‐plant resistance conditioned by Yr39 during the wheat–Puccinia striiformis f. sp tritici interaction. Mol. Plant Pathol. 9, 479–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coram, T.E. , Wang, M.N. and Chen, X.M. (2008b) Transcriptome analysis of the wheat–Puccinia striiformis f. sp tritici interaction. Mol. Plant Pathol. 9, 157–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coram, T.E. , Huang, X.L. , Zhan, G.M. , Settles, M.L. and Chen, X.M. (2010) Meta‐analysis of transcripts associated with race‐specific resistance to stripe rust in wheat demonstrates common induction of blue copper‐binding protein, heat‐stress transcription factor, pathogen‐induced WIR1A protein, and ent‐kaurene synthase transcripts. Funct. Integr. Genomics, 10, 383–392. [DOI] [PubMed] [Google Scholar]
- Cunfer, B.M. (2002) Powdery mildew. In: Bread Wheat: Improvement and Protection, FAO Plant Production and Protection Series, (Curtis B.C., Rajaram S. and Gómez Macpherson H., eds). Rome: Food and Agriculture Organization of the United Nations. Available at http://www.fao.org/icatalog/search/list_cat.asp?lang=en. [Google Scholar]
- Desmond, O.J. , Manners, J.M. , Schenk, P.M. , Maclean, D.J. and Kazan, K. (2008) Gene expression analysis of the wheat response to infection by Fusarium pseudograminearum . Physiol. Mol. Plant Pathol. 73, 40–47. [Google Scholar]
- Diethelm, M. , Rhiel, M. , Wagner, C. , Mikolajewski, S. , Groth, J. , Hartl, L. , Friedt, W. and Schweizer, G. (2011) Gene expression analysis of four WIR1‐like genes in floret tissues of European winter wheat after challenge with G. zeae . Euphytica, DOI 10.1007/s10681‐011‐0498‐7. [Google Scholar]
- Douchkov, D. , Johrde, A. , Nowara, D. , Himmelbach, A. , Lueck, S. , Niks, R. and Schweizer, P. (2011) Convergent evidence for a role of WIR1 proteins during the interaction of barley with the powdery mildew fungus Blumeria graminis . J. Plant Physiol. 168, 20–29. [DOI] [PubMed] [Google Scholar]
- Eichmann, R. , Bischof, M. , Weis, C. , Shaw, J. , Lacomme, C. , Schweizer, P. , Duchkov, D. , Hensel, G. , Kumlehn, J. and Hückelhoven, R. (2010) BAX INHIBITOR‐1 is required for full susceptibility of barley to powdery mildew. Mol. Plant–Microbe Interact. 23, 1217–1227. [DOI] [PubMed] [Google Scholar]
- Eisen, M.B. , Spellman, P.T. , Brown, P.O. and Botstein, D. (1998) Cluster analysis and display of genome‐wide expression patterns. Proc. Natl. Acad. Sci. USA, 95, 14 863–14 868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O. , Brunak, S. , von Heijne, G. and Nielsen, H. (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971. [DOI] [PubMed] [Google Scholar]
- Franck, S. and Dudler, R. (1995) Nucleotide sequence (Gen‐Bank/EMBL/DDBJ accession number X87686) of a wheat cDNA encoding a putative pathogen‐inducible protein homologous to PWIR1. Plant Physiol. 109, 338. [Google Scholar]
- Gaupels, F. , Buhtz, A. , Knauer, T. , Deshmukh, S. , Waller, F. , van Bel, A.J.E. , Kogel, K.H. and Kehr, J . (2008) Adaptation of aphid stylectomy for analyses of proteins and mRNAs in barley phloem sap. J. Exp. Bot. 59, 3297–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein, I. , Pacak, M.B. , Hrubikova, K. , Williamson, S. , Dinesen, M. , Soenderby, I.E. , Sundar, S. , Jarmolowski, A. , Shirasu, K. and Lacomme, C. (2005) Virus‐induced gene silencing‐based functional characterization of genes associated with powdery mildew resistance in barley. Plant Physiol. 138, 2155–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzberg, S. , Brosio, P. , Gross, C. and Pogue, G.P. (2002) Barley stripe mosaic virus‐induced gene silencing in a monocot plant. Plant J. 30, 315–327. [DOI] [PubMed] [Google Scholar]
- Irizarry, R.A. , Hobbs, B. , Collin, F. , Beazer‐Barclay, Y.D. , Antonellis, K.J. , Scherf, U. and Speed, T.P. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics, 4, 249–264. [DOI] [PubMed] [Google Scholar]
- Jansen, C. , Korell, M. , Eckey, C. , Biedenkopf, D. and Kogel, K.H. (2005) Identification and transcriptional analysis of powdery mildew‐induced barley genes. Plant Sci. 168, 373–380. [DOI] [PubMed] [Google Scholar]
- Jia, H.Y. , Cho, S.H. and Muehlbauer, G.J. (2009) Transcriptome analysis of a wheat near‐isogenic line pair carrying Fusarium Head Blight‐resistant and ‐susceptible alleles. Mol. Plant–Microbe Interact. 22, 1366–1378. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kankanala, P. , Czymmek, K. and Valent, B. (2007) Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell, 19, 706–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosina, P. , Reynolds, M. , Dixon, J. and Joshi, A. (2007) Stakeholder perception of wheat production constraints, capacity building needs, and research partnerships in developing countries. Euphytica, 157, 475–483. [Google Scholar]
- Krattinger, S.G. , Lagudah, E.S. , Spielmeyer, W. , Singh, R.P. , Huerta‐Espino, J. , McFadden, H. , Bossolini, E. , Selter, L.L. and Keller, B. (2009) A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science, 323, 1360–1363. [DOI] [PubMed] [Google Scholar]
- Kumar, S. , Nei, M. , Dudley, J. and Tamura, K. (2008) MEGA: a biologist‐centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9, 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löytynoja, A. and Goldman, N. (2008) Phylogeny‐aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science, 320, 1632–1635. [DOI] [PubMed] [Google Scholar]
- Marcel, T.C. , Varshney, R.K. , Barbieri, M. , Jafary, H. , de Kock, M.J.D. , Graner, A. and Niks, R.E. (2007) A high‐density consensus map of barley to compare the distribution of QTLs for partial resistance to Puccinia hordei and of defence gene homologues. Theor. Appl. Genet. 114, 487–500. [DOI] [PubMed] [Google Scholar]
- Martins‐Lopes, P. , Zhang, H. and Koebner, R. (2001) Detection of single nucleotide mutations in wheat using single strand conformation polymorphism gels. Plant Mol. Biol. Rep. 19, 159–162. [Google Scholar]
- Mauch, F. , Reimmann, C. , Freydl, E. , Schaffrath, U. and Dudler, R. (1998) Characterization of the rice pathogen‐related protein Rirla and regulation of the corresponding gene. Plant Mol. Biol. 38, 577–586. [DOI] [PubMed] [Google Scholar]
- Mellersh, D.G. and Heath, M.C. (2001) Plasma membrane–cell wall adhesion is required for expression of plant defense responses during fungal penetration. Plant Cell, 13, 413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu, C. , Keller, B. and Feuillet, C. (2003) Cytological and molecular analysis of the Hordeum vulgare–Puccinia triticina nonhost interaction. Mol. Plant–Microbe Interact. 16, 626–633. [DOI] [PubMed] [Google Scholar]
- Pogue, G.P. , Lindbo, J.A. , Dawson, W.O. and Turpen, T.H. (1998) Tobamovirus transient expression vectors: tools for plant biology and high‐level expression of foreign proteins in plants In: Plant Molecular Biology Manual (Gelvin S.B.A. and Schilperoot R.A., eds), pp. 1–27. Dordrecht: Kluwer Academic Publishers. [Google Scholar]
- Sardesai, N. , Subramanyam, S. , Nemacheck, J. and Williams, C. (2005) Modulation of defense‐response gene expression in wheat during Hessian fly larval feeding. J. Plant Interact. 1, 39–50. [Google Scholar]
- Schaffrath, U. , Mauch, F. , Freydl, E. , Schweizer, P. and Dudler, R. (2000) Constitutive expression of the defense‐related Rir1b gene in transgenic rice plants confers enhanced resistance to the rice blast fungus Magnaporthe grisea . Plant Mol. Biol. 43, 59–66. [DOI] [PubMed] [Google Scholar]
- Schreiber, A.W. , Sutton, T. , Caldo, R.A. , Kalashyan, E. , Lovell, B. , Mayo, G. , Muehlbauer, G.J. , Druka, A. , Waugh, R. , Wise, R.P. , Langridge, P. and Baumann, U. (2009) Comparative transcriptomics in the Triticeae . BMC Genomics, 10, 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer, P. , Hunziker, W. and Mosinger, E. (1989) cDNA cloning, in vitro transcription and partial sequence‐analysis of messenger‐RNAs from winter‐wheat (Triticum‐aestivum L) with induced resistance to Erysiphe‐graminis f‐sp tritici . Plant Mol. Biol. 12, 643–654. [DOI] [PubMed] [Google Scholar]
- Schweizer, P. , Buchala, A. , Dudler, R. and Metraux, J.P. (1998) Induced systemic resistance in wounded rice plants. Plant J. 14, 475–481. [Google Scholar]
- Schweizer, P. , Pokorny, J. , Abderhalden, O. and Dudler, R.A. (1999) A transient assay system for the functional assessment of defence‐related genes in wheat. Mol. Plant–Microbe Interact. 12, 647–654. [Google Scholar]
- Scofield, S.R. , Huang, L. , Brandt, A.S. and Gill, B.S. (2005) Development of a virus‐induced gene‐silencing system for hexaploid wheat and its use in functional analysis of the Lr21‐mediated leaf rust resistance pathway. Plant Physiol. 138, 2165–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R.P. , Huerta‐Espino, J. , Bhavani, S. , Herrera‐Foessel, S.A. , Singh, D. , Singh, P.K. , Velu, G. , Mason, R.E. , Jin, Y. and Crossa, J. (2011) Race non‐specific resistance to rust diseases in CIMMYT spring wheats. Euphytica, 179, 175–186. [Google Scholar]
- Smyth, G.K. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article 3. [DOI] [PubMed] [Google Scholar]
- Tufan, H.A. , McGrann, G.R.D. , Magusin, A. , Morel, J.B. , Miche, L. and Boyd, L.A. (2009) Wheat blast: histopathology and transcriptome reprogramming in response to adapted and nonadapted Magnaporthe isolates. New Phytol. 184, 473–484. [DOI] [PubMed] [Google Scholar]
- Tufan, H.A. , Stefanato, F.L. , McGrann, G.R.D. , MacCormack, R. and Boyd, L.A. (2011) The Barley stripe mosaic virus system used for virus‐induced gene silencing in cereals differentially affects susceptibility to fungal pathogens in wheat. J. Plant Physiol. 168, 990–994. [DOI] [PubMed] [Google Scholar]
- Urashima, A.S. , Igarashi, S. and Kato, H. (1993) Host range, mating type, and fertility of Pyricularia grisea from wheat in Brazil. Plant Dis. 77, 1211–1216. [Google Scholar]
- Urashima, A.S. , Lavorent, N.A. , Goulart, A.C.P. and Mehta, Y.R. (2004) Resistance spectra of wheat cultivars and virulence diversity of Magnaporthe grisea isolates in Brazil. Fitopatol. Bras. 29, 511–518. [Google Scholar]
- Vandesompele, J. , De Preter, K. , Pattyn, F. , Poppe, B. , Van Roy, N. , De Paepe, A. and Speleman, F. (2002) Accurate normalization of real‐time quantitative RT‐PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, research0034.0031–research0034.0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wäspi, U. , Blanc, D. , Winkler, T. , Ruedi, P. and Dudler, R. (1998) Syringolin, a novel peptide elicitor from Pseudomonas syringae pv. syringae that induces resistance to Pyricularia oryzae in rice. Mol. Plant–Microbe Interact. 11, 727–733. [Google Scholar]
- Welham, S.J. (1993) Procedure GLMM In: GenStat 5 Procedure Library Manual Release 3 (Payne R.W., Arnold G.M. and Morgan G.W., eds), pp. 187–192. Oxford: Numerical Algorithms Group. [Google Scholar]
- Wettenhall, J.M. , Simpson, K.M. , Satterley, K. and Smyth, G.K. (2006) affylmGUI: a graphical user interface for linear modeling of single channel microarray data. Bioinformatics, 22, 897–899. [DOI] [PubMed] [Google Scholar]
- Wise, R.P. , Caldo, R.A. , Hong, L. , Shen, L. , Cannon, E. and Dickerson, J.A. (2007) BarleyBase/PLEXdb: a unified expression profiling database for plants and plant pathogens In: Methods in Molecular Biology: Plant Bioinformatics—Methods and Protocols (Edwards D., ed.), pp. 347–363. Totowa, NJ: Humana Press. [Google Scholar]
- Yuan, H.Y. , Chen, X.P. , Zhu, L. and He, G.C. (2004) Isolation and characterization of a novel rice gene encoding a putative insect‐inducible protein homologous to wheat Wir1 . J. Plant Physiol. 161, 79–85. [DOI] [PubMed] [Google Scholar]
- Zellerhoff, N. , Himmelbach, A. , Dong, W.B. , Bieri, S. , Schaffrath, U. and Schweizer, P. (2010) Nonhost resistance of barley to different fungal pathogens is associated with largely distinct, quantitative transcriptional responses. Plant Physiol. 152, 2053–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierold, U. , Scholz, U. and Schweizer, P. (2005) Transcriptome analysis of mlo‐mediated resistance in the epidermis of barley. Mol. Plant Pathol. 6, 139–151. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. (2009) Early molecular events in PAMP‐triggered immunity. Curr. Opin. Plant Biol. 12, 414–420. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 CLUSTALW nucleotide alignment of BSMV:WIR1 and TaWIR1 sequences showing conservation of the target region between TaWIR1 genes. Shading indicates 100% (black), 75% (dark grey) and 50% (light grey) conservation between sequences.
Fig. S2 Barley stripe mosaic virus (BSMV)‐mediated virus‐induced gene silencing (VIGS) of TaWIR1 reduces the expression of transcripts corresponding to probe sets Ta.21556.1, Ta.22732.1 and Ta.3133.1 on the Affymetrix Wheat GeneChip, despite these sequence having less than 80% similarity to BSMV:WIR1. Transcript levels were measured by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) and shown relative to mock‐inoculated controls. The mean values of three independent experiments are shown with standard error bars.
Fig. S3 Transcript levels of TaWIR1 genes in intact leaves of cv. Renan measured after inoculation (hours post‐inoculation, hpi) with Magnaporthe oryzae isolate BR32, relative to mock‐inoculated controls. The mean values of two independent experiments are shown with standard error bars.
Fig. S4 Transcript levels of TaWIR1 genes in cv. Renan leaf tissues inoculated with Magnaporthe oryzae isolate BR32, relative to the same region of the leaf in the mock‐inoculated control. The mean values of three independent experiments are shown with standard error bars.
Table S1 TaWIR1 transcripts present as probe sets on the Affymetrix Wheat GeneChip.
Table S2 Primer sequences.
Table S3 TaWIR1 genes identified from database similarity searches.
Table S4 SignalP analysis of TaWIR1 proteins.
Table S5 TargetP analysis of TaWIR1 proteins.
Table S6 Transmembrane prediction for TaWIR1 proteins.
Table S7 Nucleotide identities of TaWIR1 sequences to BSMV:WIR1.
Table S8 BSMV:WIR1 silencing targets predicted by si‐Fi analysis.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item


