Figure 1.
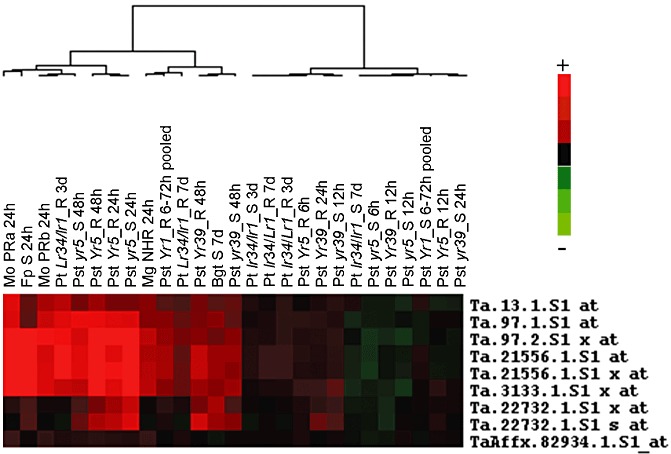
Meta‐analysis of TaWIR1 probe sets on the Affymetrix Wheat GeneChip differentially transcribed in response to fungal pathogen inoculation. Experiment descriptions are shown at the top of the figure, with the probe set names listed on the right of the figure. Experiments included are Magnaporthe oryzae (Mo) inoculations with isolate BR32 (a) or BR37 (b), partial resistance (PR) interactions, with M. grisea (Mg) isolate BR29, a nonhost interaction (NHR) (Tufan et al., 2009), incompatible and compatible interactions with Puccinia striiformis f. sp. tritici (Pst) (Yr5, Yr39, Yr1; 2008a, 2008b and Bozkurt et al., 2010, respectively), incompatible and compatible interactions with P. triticina (Pt) (Lr34 and Lr1; Bolton et al., 2008), and compatible Blumeria graminis f. sp. tritici (Bgt; Chain et al., 2009) and Fusarium pseudograminearum (Fp; Desmond et al., 2008) inoculations. The time points sampled post‐inoculation are shown alongside the pathogen labels. Red blocks indicate transcript up‐regulation and green blocks indicate transcript down‐regulation in pathogen‐inoculated tissue relative to controls. Samples represent leaf‐inoculated tissues, except Fp, which is from inoculated stem bases.
