Figure 1.
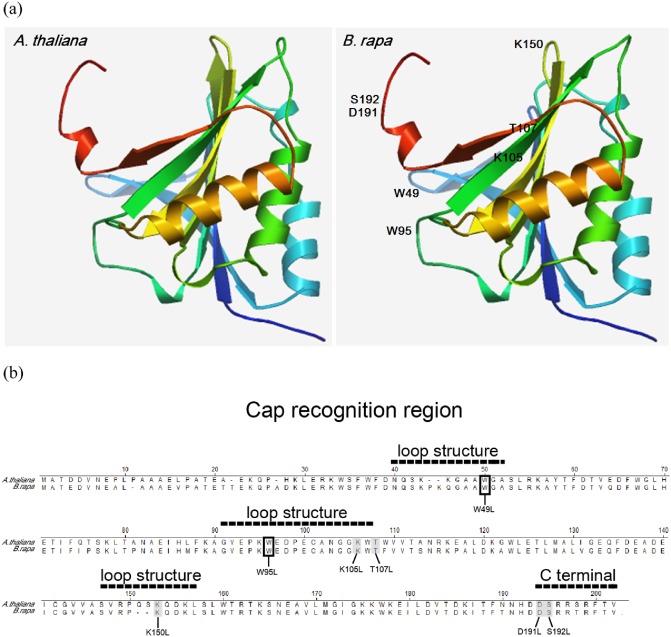
Positions of the mutated amino acids in the cap‐binding pocket of eIF(iso)4E [eukaryotic initiation factor(iso)4E]. (a) Comparison of predicted three‐dimensional structural models of Arabidopsis thaliana eIF(iso)4E and Brassica rapa eIF(iso)4E. The structures are shown in a ribbon representation. Amino acids of B. rapa eIF(iso)4E mutated in this work are labelled. The X‐ray crystal structure of wheat eIF4E (Protein Data Bank accession no. 2IDR) was used as the template structure. (b) Alignment of A. thaliana and B. rapa eIF(iso)4E amino acid sequences. Seven candidate cap‐binding pocket amino acids (Trp49, Trp95, Lys105, Thr107, Lys150, Asp191 and Ser192) from B. rapa that were mutated are indicated with grey boxes in the sequence alignment.
