SUMMARY
Pseudomonas syringae pv. actinidiae is the causal agent of bacterial canker of green‐fleshed kiwifruit (Actinidia deliciosa) and yellow‐fleshed kiwifruit (A. chinensis). A recent, sudden, re‐emerging wave of this disease has occurred, almost contemporaneously, in all of the main areas of kiwifruit production in the world, suggesting that it can be considered as a pandemic disease. Recent in‐depth genetic studies performed on P. syringae pv. actinidiae strains have revealed that this pathovar is composed of four genetically different populations which, to different extents, can infect crops of the genus Actinidia worldwide. Genome comparisons of these strains have revealed that this pathovar can gain and lose the phaseolotoxin gene cluster, as well as mobile genetic elements, such as plasmids and putative prophages, and that it can modify the repertoire of the effector gene arrays. In addition, the strains currently causing worldwide severe economic losses display an extensive set of genes related to the ecological fitness of the bacterium in planta, such as copper and antibiotic resistance genes, multiple siderophore genes and genes involved in the degradation of lignin derivatives and other phenolics. This pathogen can therefore easily colonize hosts throughout the year.
Taxonomy: Bacteria; Proteobacteria, gamma subdivision; Order Pseudomonadales; Family Pseudomonadaceae; Genus Pseudomonas; Pseudomonas syringae species complex, genomospecies 8; Pathovar actinidiae.
Microbiological properties: Gram‐negative, aerobic, motile, rod‐shaped, polar flagella, oxidase‐negative, arginine dihydrolase‐negative, DNA 58.5–58.8 mol.% GC, elicits the hypersensitive response on tobacco leaves.
Host range: Primarily studied as the causal agent of bacterial canker of green‐fleshed kiwifruit (Actinidia deliciosa), it has also been isolated from yellow‐fleshed kiwifruit (A. chinensis). In both species, it causes severe economic losses worldwide. It has also been isolated from wild A. arguta and A. kolomikta.
Disease symptoms: In green‐fleshed and yellow‐fleshed kiwifruits, the symptoms include brown–black leaf spots often surrounded by a chlorotic margin, blossom necrosis, extensive twig die‐back, reddening of the lenticels, extensive cankers along the main trunk and leader, and bleeding cankers on the trunk and the leader with a whitish to orange ooze.
Epidemiology: Pseudomonas syringae pv. actinidiae can effectively colonize its host plants throughout the year. Bacterial exudates can disperse a large amount of inoculum within and between orchards. In the spring, temperatures ranging from 12 to 18 °C, together with humid conditions, can greatly favour the multiplication of the bacterium, allowing it to systemically move from the leaf to the young shoots. During the summer, very high temperatures can reduce the multiplication and dispersal of the bacterium. Some agronomical techniques, as well as frost, wind, rain and hail storms, can contribute to further spreading.
Disease control: An integrated approach that takes into consideration precise scheduled spray treatments with effective and environmentally friendly bactericides and equilibrated plant nutrition, coupled with preventive measures aimed at drastically reducing the bacterial inoculum, currently seems to be the possible best solution for coexistence with the disease. The development of resistant cultivars and pollinators, effective biocontrol agents, including bacteriophages, and compounds that induce the systemic activation of plant defence mechanisms is in progress.
Useful websites: Up‐to‐date information on bacterial canker research progress and on the spread of the disease in New Zealand can be found at: http://www.kvh.org.nz. Daily information on the spread of the disease and on the research being performed worldwide can be found at: http://www.freshplaza.it.
INTRODUCTION
Pseudomonas syringae pv. actinidiae is the causal agent of bacterial canker of green‐fleshed kiwifruit (Actinidia deliciosa) and yellow‐fleshed kiwifruit (A. chinensis), and is currently the cause of severe economic losses in countries such as Italy and New Zealand, where kiwifruit is a major crop. In addition, the disease also causes important economic losses in France, Spain, Portugal, Chile, South Korea and Japan. The bacterial canker of kiwifruit incited by P. s. pv. actinidiae fulfils all of the definitions for an emerging or re‐emerging infectious disease: (i) it has increased in incidence, and expanded its geographical or host range; (ii) it has exhibited a changed or modified pathogenesis; (iii) it is newly evolved; and (iv) it is newly recognized (Anderson et al., 2004; Daszak et al., 2000). Among these main drivers of the emergence of bacterial plant diseases, the introduction of the pathogen into a new area and the occurrence of particular weather conditions favourable to the pathogen are seen as fundamental (Anderson et al., 2004). Indeed, both factors have played a main role in the recent epidemics of bacterial canker of kiwifruit.
Pseudomonas syringae pv. actinidiae was first isolated, identified and described in Japan in 1984 (Takikawa et al., 1989). Subsequently, it has been found in Italy and South Korea (Koh et al., 1994; Scortichini, 1994). In all cases, the pathogen was isolated from A. deliciosa cv. Hayward. Interestingly, although the disease was destructive in Japan and South Korea, in Italy, over a period of 20 years on about 24 000 hectares of A. deliciosa spread all over the peninsula, P. s. pv. actinidiae did not cause significant losses. However, during 2008–2011, sudden and repeated epidemics of bacterial canker posed a serious threat to the cultivation of the highly prized yellow‐fleshed kiwifruit in central Italy, where almost all of the 800 hectares devoted to this crop were heavily infected. Consequently, most of the orchards were uprooted because of the very high susceptibility of the A. chinensis germplasm currently cultivated (Hort16A, Jin Tao, Soreli) (Balestra et al., 2009; Ferrante and Scortichini, 2009, 2010).
During 2009–2011, P. s. pv. actinidiae also started to severely infect A. deliciosa cv. Hayward in all of the main producing regions in Italy. In 2010, the pathogen was also officially reported in New Zealand, where, during 2011, it caused very severe damage to both A. chinensis and A. deliciosa (Everett et al., 2011; http://www.kvh.org.nz). Recently, the pathogen has also been recorded for the first time in Portugal, France, Spain, Switzerland, Turkey and Chile (Abelleira et al., 2011; Balestra et al., 2010; Bastas and Karakaya, 2012; European Plant Protection Organization, 2011a, 2011b; Vanneste et al., 2011a), and it has been reported again in South Korea (Koh et al., 2010). In some countries (i.e. Portugal, Spain, France, South Korea), the bacterium has infected both green‐fleshed and yellow‐fleshed kiwifruits. Remarkably, during the last 20 years, P. s. pv. actinidiae has also been isolated from both A. chinensis and A. deliciosa in China (Liang et al., 2000; Wang et al., 1992). The almost contemporaneous presence of the same very aggressive pathogen in all areas of the world in which kiwifruits are grown allows us to define the current repeated outbreaks of bacterial canker in A. deliciosa and A. chinensis as a real pandemic disease. Recent in‐depth studies on P. s. pv. actinidiae genomes have shown that this pathovar can rapidly adapt to a new host and new environments through the acquisition and/or loss of mobile genetic elements and virulence factors, thereby resulting in a multi‐faceted plant pathogen (Marcelletti et al., 2011).
In this review, we focus concisely on the various aspects of taxonomy, population structure, pathogen origin, detection, epidemiology, pathogenic evolution and ecological fitness of P. s. pv. actinidiae. Knowledge about these issues is considered to be very important for the development of integrated strategies aimed at reducing further the spread of this pathogen within and between orchards and countries.
TAXONOMY
On the basis of field symptoms and host range tests, and with the aid of biochemical, physiological, nutritional tests and molecular typing, P. syringae (i.e. the P. syringae species complex) has been divided into 57 pathovars (Bull et al., 2010). To genetically describe 48 P. syringae pathovars and some related species of the phytopathogenic pseudomonads, Gardan et al. (1999) performed DNA–DNA hybridization and ribotyping analyses and described nine discrete genomospecies. However, P. s. pv. actinidiae was not included in this study. By performing biochemical tests, repetitive‐sequence polymerase chain reaction (PCR), amplified ribosomal DNA restriction analysis (ARDRA) and amplified fragment length polymorphism (AFLP) analyses, this pathovar was subsequently placed into genomospecies 8, together with P. avellanae and P. s. pv. theae (Manceau and Brin, 2003; Scortichini et al., 2002).
Multilocus sequence typing (MLST) analysis confirmed these data and revealed that P. s. pv. actinidiae and P. s. pv. theae are more closely related to each other than to P. avellanae (Marcelletti et al., 2011). This was shown in a phylogenetic comparison among the other genomospecies of the P. syringae complex in a dendrogram constructed by the neighbour‐joining (NJ) algorithm, using concatenated gyrB, rpoB and rpoD gene fragments for a total of 1646 nucleotides, and in a maximum likelihood (ML) phylogram based on the acnB, fruK, gltA, pgi, rpoB and rpoD gene fragments, for a total of 2926 nucleotides.
POPULATION STRUCTURE
One of the most striking results recently highlighted by in‐depth genetic studies is the occurrence of several clearly different populations of the pathovar actinidiae which are able to infect Actinidia species to different extents. Indeed, there are presently at least four genetically distinct P. s. pv. actinidiae populations spread in different areas of the world. This represents a remarkable case of convergent evolution of genetically different populations to the same plant genus.
The first population comprises strains associated with the initial epidemics of bacterial canker observed in Japan (1984–1989) and Italy (1992), isolated in both countries from A. deliciosa cv. Hayward. The strains of this population are almost identical genetically but, interestingly, they caused severe economic losses in Japan, with only sporadic and minor damage in Italy during a period of almost 20 years (Ferrante and Scortichini, 2010; Marcelletti et al., 2011; Takikawa et al., 1989). This remarkably diverse behaviour clearly suggests that different climatic parameters and/or agronomical techniques can play a fundamental role in determining the virulence of the same bacterial pathogen. All strains of this population possess the phaseolotoxin gene cluster located in the chromosome, acquired by horizontal gene transfer (1997, 1999).
The second population has only been isolated in South Korea and is characterized by the presence of a plasmid‐borne gene that putatively produces coronatine and by the absence of the phaseolotoxin gene cluster (Han et al., 2003a). MLST analysis has revealed that this population is divergent from the first (Chapman et al., 2011). In South Korea, this population has caused severe economic losses to A. deliciosa cv. Hayward in both the past and present, as well as to A. chinensis Hort16A in recent epidemics (1994, 2010). After a few years of spray treatments, strains of these first two populations developed resistance to both copper and streptomycin (Goto et al., 1994; 2003b, 2004; 1995, 2002).
The third population is the pandemic form, currently causing very severe economic losses worldwide. In New Zealand, it was named first as ‘Italian’ and afterwards as Psa‐V (i.e. virulent). Molecular typing, MLST analysis, genome sequencing and pathogenicity tests have revealed that this population is different from the other two (Chapman et al., 2011; Ferrante and Scortichini, 2010, 2011; Gallelli et al., 2011a;, Marcelletti et al., 2011). In addition, the strains belonging to this population lack genes for coronatine and phaseolotoxin production. After its first appearance in 2008 in the Latium region (i.e. central Italy), it subsequently (i.e. 2–3 years) spread to all of the main Italian areas of kiwifruit cultivation. MLST analysis, coupled with linkage disequilibrium tests, has indicated that, in Italy, this population is currently clonal, and apparently has totally replaced the previous population (Marcelletti and Scortichini, 2011). In New Zealand, this population is currently affecting orchards in the main areas of production of kiwifruit. This population also displays identical molecular features in New Zealand, France, Spain and Portugal (Abelleira et al., 2011; Balestra et al., 2010; Chapman et al., 2011; Vanneste et al., 2011a).
The fourth population is the so‐called Psa‐LV form (i.e. less virulent), which has only been found in very few orchards in New Zealand, apparently causing only leaf spot symptoms. Interestingly, this population has been recorded only in the South Island (http://www.kvh.org.nz; 9 November 2011). MLST and effector analyses have clearly shown that this P. s. pv. actinidiae population is diverse from the other three (Chapman et al., 2011). An apparently similar P. s. pv. actinidiae strain, characterized by low virulence, has also been reported in Australia (Australian Government, Biosecurity Australia, 2011)
PATHOGEN ORIGIN AND EVOLUTIONARY ASPECTS
One of the most intriguing research objectives of the sudden and destructive recent pandemic of bacterial canker of kiwifruit worldwide has been to determine the possible origin of this very aggressive P. s. pv. actinidiae population and to elucidate some of the evolutionary aspects that are present in the different populations of this pathovar. An in‐depth assessment and comparison of the open reading frames (ORFs) of three representative P. s. pv. actinidiae strains (i.e. the type strain of the pathovar isolated in Japan in 1984, one strain isolated 20 years ago and another strain isolated from the recent epidemics in Italy) and related species and pathovars have been performed recently (Marcelletti et al., 2011).
In this study, 35 Pseudomonas spp. and pathovar genomes that were available as drafts or as complete genome sequences were compared. The results showed that, of the 398 ORFs specific for the recent very aggressive P. s. pv. actinidiae population, there were 238 ORFs for which no homologue could be found in P. avellanae or in P. s. pv. theae type strains. In addition, it was found that 49% of these 398 proteins that did not have homologous matches in the P. s. pv. actinidiae strains of past outbreaks in Japan and Italy did have matched sequences in at least one of the genomes of the P. s. pv. tomato strains examined. This evidence, together with the annotation of several deduced proteins that matched phage or prophage proteins, strongly suggests that a large portion of this specific DNA genome has been acquired by the very aggressive population through horizontal gene transfer.
Furthermore, 171 ORFs from the list of genes that were found to be conserved among P. s. pv. actinidiae strains and in P. avellanae and P. s. pv. theae type strains were selected, concatenated and aligned to determine the genealogy with the ML algorithm and to perform hypothesis testing. On the basis of these results (Fig. 1a), and on the statistical support for the rejection of the hypotheses of alternative evolutionary scenarios (Fig. 1b), the P. s. pv. actinidiae strain of the recent epidemics in Italy clearly originated from a common ancestor of the P. s. pv. actinidiae strains of past epidemics in Japan and Italy, and was not a derivative of these strains.
Figure 1.
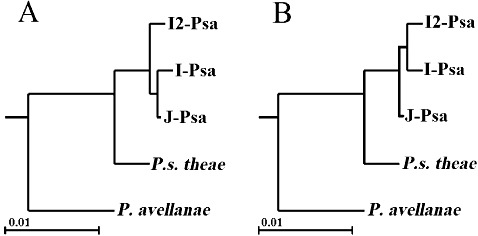
Genealogy of Pseudomonas strains of genomospecies 8 sensu Gardan et al. (1999): P. avellanae, P. syringae pv. theae and P. s. pv. actinidiae. (A) A maximum likelihood tree resulting from the analysis of the concatenation of 171 open reading frames (ORFs). (B) An alternative genealogical hypothesis with the constraint of the common origin of the Italian P. s. pv. actinidiae strain that was rejected by the monophyly test. Psa, P. s. pv. actinidiae; J, NCPPB3739 from Japan, isolated in 1984; I, NCPPB3871 from Italy, isolated in 1992; I2, CRA‐FRU 8.43 from Italy, isolated in 2008. The National Collection of Plant Pathogenic Bacteria (NCPPB) strains are representative of past outbreaks, whereas CRA‐FRU 8.43 has been isolated in the recent pandemic. [From Marcelletti et al. (2011).]
As a further confirmation of this view, within the group of 398 ORFs specific for the strains of the current very aggressive P. s. pv. actinidiae population, eight ORFs that showed significant blastn matches to sequences in the draft genomes of P. avellanae or P. s. pv. theae type strains, but did not match any of the other 35 Pseudomonas spp. and pathovar genome sequences, were detected. The most obvious explanation for this result is that these eight sequences were shared by P. s. pv. actinidiae strains of the current epidemics, and other strains of genomospecies 8 were lost by the P. s. pv. actinidiae strains of the past epidemics in Japan and Italy during their evolution. All of the results congruently suggest that the very aggressive population that has currently spread to different continents did not evolve from the organisms that caused the epidemics in 1984–1992 in Japan and Italy, but rather from a common ancestor.
Taking into account that P. s. pv. actinidiae was isolated from wild Actinidia species (i.e. A. arguta, A. kolomikta) in Japan (Ushiyama et al., 1992a), and that the Actinidiaceae family originated in eastern Asia, it is also conceivable that P. s. pv. actinidiae originated in eastern Asia, as has been proposed by Ushiyama et al. (1992b).
As far as the rapid spread of the pathogen on a large scale is concerned, evidence in Italy suggests that the distribution of latently infected propagative material has largely contributed to the dissemination of P. s. pv. actinidiae all over the country. It remains to be established from where the pathogen arrived in Italy, because genetic studies have demonstrated that the pandemic population did not evolve from the previous population that was already present. The introduction of latently infected plant material has also been implicated as the cause for the occurrence of kiwifruit bacterial canker in France, Spain, Portugal, Switzerland and Chile. In New Zealand, anecdotal evidence points to latently infected pollen imported from abroad as the main cause of the epidemics of bacterial canker.
DETECTION
The isolation, detection and presumptive identification of putative P. s. pv. actinidiae colonies grown on bacterial culture medium are not difficult. A two‐step PCR‐based method can yield reliable results. A preliminary screening of the pure colonies can be carried out by applying the PCR techniques described by Koh and Nou (2002) or Rees‐George et al. (2010). Both procedures appear to be satisfactory for a preliminary investigation, although sometimes aspecific bands and the amplification from P. s. pv. theae can be obtained with both techniques (Gallelli et al., 2011a). Colony identity can be confirmed by repetitive‐PCR fingerprinting (i.e. BOX‐PCR and/or ERIC‐PCR) (Fig. 2a,b) (Ferrante and Scortichini, 2009, 2010).
Figure 2.
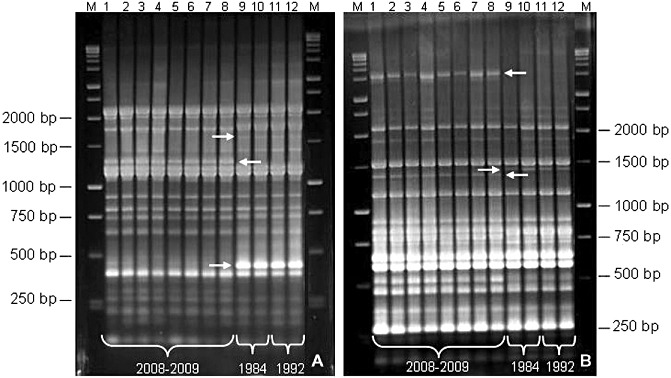
Repetitive‐sequence polymerase chain reaction (PCR) (A, BOX‐PCR; B, ERIC‐PCR) fingerprint patterns for genomic DNAs of Pseudomonas syringae pv. actinidiae strains isolated in central Italy from Actinidia chinensis and A. deliciosa during 2008–2009, compared with strains of past outbreaks of bacterial canker of kiwifruit in Japan (1984) and Italy (1992). [From Ferrante and Scortichini (2010).] The arrows indicate the bands enabling the visible differentiation of the two populations.
The rapid and reliable detection of propagative material of kiwifruit circulating worldwide is a fundamental step toward reducing the further risk of spread of the pathogen within and between countries. A technique that determines the presence of the pathogen directly in different plant organs, including pollen, has been developed recently by Gallelli et al. (2011a). Accordingly, a duplex‐PCR based on the primers of Koh and Nou (2002) and on the avrD1 gene has enabled the detection of the pathogen directly from artificially contaminated bark, leaves, flowers, petioles and pollen, without prior isolation on bacterial culture medium. When applied to naturally infected kiwifruit material, the procedure also yields good results. The technique may be applied for the specific identification of colonies, avoiding false positive results caused by the genetic similarity with P. s. pv. theae. The reliability when applied to the apparently healthy propagative kiwifruit material remains to be verified, although the reported sensitivity [i.e. 2 × 10 colony‐forming units (cfu)/PCR] is excellent.
PATHOGENIC EVOLUTION
Pseudomonas syringae pv. actinidiae is a vivid example of how plant pathogenic bacteria can rapidly evolve and modulate their pathogenicity and virulence to adapt to new host plants and environments. All of the four populations described above are able to incite clear disease symptoms in Actinidia species, even though some strains of the same population do not display all of their pathogenic potential in every circumstance. In the present scenario, the occurrence or absence of certain virulence factors, such as the phytotoxins coronatine and phaseolotoxin, does not seem to play a relevant role in enhancing the disease symptoms.
Therefore, the question that arises is how did this new highly virulent P. s. pv. actinidiae population originate? The importance of stress factors in the promotion of bacterial evolution has been emphasized recently. Under stress conditions in the host (i.e. nutrient deficiency outside the host, attack by antimicrobial compounds inside the host and low temperatures), the bacterial competency for DNA uptake is activated and the pathogen can acquire exogenous genetic material that could help it to escape from the stress (Arnold et al., 2007). In addition, a possible loss of mobile genetic elements carrying avirulence genes can lead to enhanced virulence (Jackson et al., 2011a). Genome comparisons have revealed that the phaseolotoxin gene cluster, as well as plasmids and effector genes, can be gained and lost by P. s. pv. actinidiae strains. In addition, horizontal gene transfer has played a relevant role in shaping the genomes of the strains of the current pandemic population (Marcelletti et al., 2011). Among the stress conditions that may have putatively promoted the outbreak and spread of the pathogen, it should be pointed out that the winter and spring frosts that occurred during the winter of 2007–2008 in Latium (i.e. central Italy) may have been a factor. This may be particularly significant given that A. chinensis originated from subtropical areas of eastern China. Latent cells of the bacterium that were already present in the plant could have been favoured for their further colonization by the presence of damaged tissues that occurred as a result of the low temperatures (i.e. until −9 °C).
An assessment of the presence of the 12 effectors in P. s. pv. actinidiae strains of the recent pandemic in Italy and New Zealand did not reveal any differences between the strains (Chapman et al., 2011). In addition, a comparison of the effector repertoire of three P. s. pv. actinidiae strains based on genome analysis revealed a ‘core’ set of 33 hop and six avr putative effector genes that were conserved in the three strains. In general, in these putative effectors, the amino acid identity to the most similar orthologue of the P. syringae pathovars found in GenBank was very high (i.e. >90%) (Marcelletti et al., 2011). Interestingly, the current, very aggressive population displays four putative effector genes, namely hopA1, hopAA1‐2, hopH 1 and hopZ2‐like, which were not present in the strains isolated in Japan and Italy 20–28 years ago (Fig. 3). These results indicate that both A. deliciosa and A. chinensis can be infected by different P. s. pv. actinidiae strains displaying different repertoires of effector genes. To better clarify the evolution of the pathogenicity within P. syringae pathovars and Pseudomonas species (Arnold and Jackson, 2011; Ferrante and Scortichini, 2011), the role(s) of the specific array of effector genes found in the pandemic population must be verified.
Figure 3.
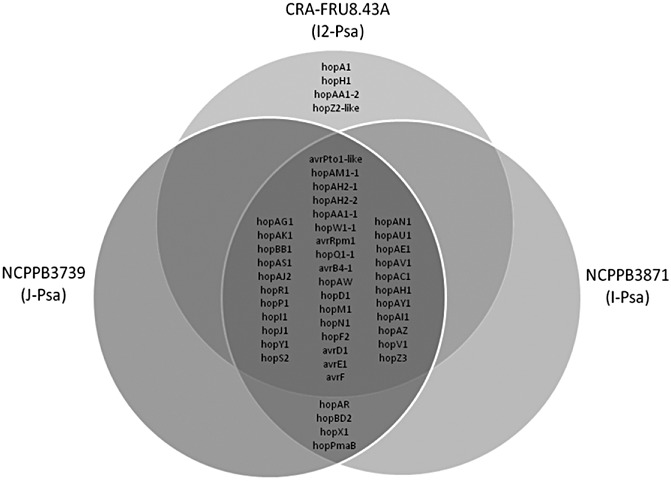
Venn diagram of the type III effector gene complements of sequenced Pseudomonas syringae pv. actinidiae strains based on the comparison of the same complement of other sequenced plant pathogenic pseudomonads. The genes conserved among the three strains are indicated in the middle of the diagram. Strains of past (NCPPB3739 from Japan and NCPPB3871 from Italy) and current (CRA‐FRU 8.43) epidemics of bacterial canker display four unique different effector genes. [From Marcelletti et al. (2011).]
The Japanese strains from the 1984 outbreak, as well as the strains found in Italy 20 years ago, share a native plasmid of about 50 kb. This plasmid is absent in the P. s. pv. actinidiae strains isolated in Italy during the recent epidemics of bacterial canker in A. chinensis and A. deliciosa. By contrast, the recently isolated strains harbour a different plasmid of about 160 kb in length. This represents a substantial acquisition of genetic material for such a new population. Interestingly, the three P. s. pv. actinidiae strains that have been sequenced possess the repA gene, which is essential for the replication of the P. syringae pPT23A‐like plasmid family. Plasmids of this family possess genes encoding virulence factors, such as type III effectors, phytotoxins, plant hormones and determinants, important for epiphytic fitness, as well as genes for conjugation and insertion sequence elements (Sundin, 2007). On a 150‐kb plasmid of this family found in P. s. pv. phaseolicola 1448A, a 30‐kb pathogenic island encoding three type III effectors and other potential virulence genes relevant for strain virulence in bean (Jackson et al., 1999) has been identified. Interestingly, Baltrus et al. (2011) revealed a virulence gene repertoire of type III effectors for another P. s. pv. actinidiae strain isolated in Japan, which was similar to the virulence gene present on the large virulent plasmid found in P. s. pv. phaseolicola 1448A. Whether or not this large plasmid also migrated into the pandemic P. s. pv. actinidiae population remains an interesting hypothesis, which requires testing.
In a variable region (i.e. DNA region larger than 10 kb in a contig that appeared as a gap in the genome alignment and genomic regions showing a different G + C content with respect to the average content of the three P. s. pv. actinidiae strains) of the strain isolated during the recent epidemics, many putative proteins were found that were related to the assembly and acquisition of prophage PSPPHO6, which were also present in P. s. pv. phaseolicola 1448A. This region is not present in the strains responsible for the past outbreaks. A direct link between the enhanced virulence or further host adaptation of the pandemic population in Actinidia spp. and the presence of this prophage, which was acquired by horizontal gene transfer, remains to be established. It is worth remembering that phages, prophages and their morons (i.e. DNA elements inserted between a pair of genes in one phage genome) are known to shape the pathogenicity and virulence of bacterial pathogens, and their presence within the bacterial genome can largely contribute to the genetic and phenotypic diversity of bacteria and to the emergence of pathogenic variants (Ronning et al., 2010, Jackson et al., 2011b). In Xylella fastidiosa, the causative agent of infectious diseases of many cultivated crops, prophage‐associated chromosomal rearrangements and deletions have been found to be largely responsible for strain‐specific differences (Van Sluys et al., 2003).
ECOLOGICAL FITNESS
The P. s. pv. actinidiae genome also includes sets of genes that are important for the survival of the bacterium or for competing with other micro‐organisms in planta. Indeed, the pathogen can inhibit host nitric oxide metabolism. Nitric oxide plays a fundamental role in plant disease resistance by acting as a signal‐inducing plant gene to synthesize defence‐related compounds. The inhibition of nitric oxide synthesis consequently promotes the bacterial growth in planta (Delledonne et al., 1998). Furthermore, P. s. pv. actinidiae genomes contain copA and copB, genes that play a key role in copper resistance. Copper ions are essential for bacterial species, but can induce toxic cellular effects if levels of free ions are not controlled (Cooksey, 1993). Interestingly, Nakajima et al. (2002) found that, at the beginning of bacterial canker outbreaks in Japan (i.e. 1984–1987), all the P. s. pv. actinidiae strains displayed only copA and copB. However, after repeated spray treatments with copper‐based bactericides, the pathogen also showed additional genes responsible for maximal resistance to copper, namely copR and copS.
Pseudomonas syringae pv. actinidiae can counteract the lethal effect of antibiotics by means of multidrug efflux pumps of the multidrug resistance systems encoded by chromosomal genes. Notably, the genome of the strain from the pandemic population also encodes a dehydratase protein which might putatively inactivate lantibiotic antibiotics, which are produced by Gram‐positive bacteria and characterized by high specific activity against multidrug‐resistant bacteria (Brotz and Sahl, 2000). It is worth noting that P. s. pv. actinidiae strains can easily tolerate streptomycin through resistance mechanisms, as already observed in Japan and South Korea (2003b, 2004; Lee et al., 2005; Nakajima et al., 1995)
The efficient uptake and utilization of iron through siderophores is regarded as an important virulence factor for phytopathogenic pseudomonads, especially in iron‐limited environments (Neilands, 1995). Pseudomonas syringae pv. actinidiae strains possess a set of genes coding for the production of siderophores, such as pyoverdine, haemin, enterobactin and yersiniabactin. The last two siderophores, primarily described in the Enterobacteriaceae, are characterized by a very high affinity for iron (Carniel, 2001; Raymond et al., 2003).
Although the P. s. pv. actinidiae strains analysed by genome sequencing were isolated from leaf spot symptoms, they display a set of genes involved in the degradation of lignin derivatives and other phenolics. Similar to other P. syringae pathovars associated with woody hosts, such as P. s. pv. aesculi and P. savastanoi pv. savastanoi (Green et al., 2010; Rodriguez‐Palenzuela et al., 2010), and to soil‐inhabiting species, such as P. putida, P. s. pv. actinidiae strains have genes putatively related to the degradation of anthranilate and protocatechuate. These pathways allow for the utilization of unsubstituted lignin‐related compounds and other plant‐derived phenolic compounds, such as mandalate and phenol.
EPIDEMIOLOGY AND FIELD VIRULENCE
The most significant aspect of bacterial canker disease of kiwifruit is, most probably, the ease with which the bacterium spreads within and between orchards. Apparently healthy plants can show the first sign of infection in the spring (i.e. leaf spots) and then die by the end of the year. Similarly, orchards with no signs of infection and that are hundreds of metres away from an infected site, can show signs of infection on most of the plants within a year.
Bacterial exudates, which ooze from the cankers during the end of autumn–winter and early spring and are dispersed by the wind, are very effective at spreading the inoculum within and between orchards (Serizawa et al., 1989; Serizawa and Ichikawa, 1993a). Springs characterized by frequent rains and/or high humidity and optimal temperatures (12–18 °C) are also indicated as very conducive for rapid pathogen multiplication (Serizawa and Ichikawa, 1993b). During the spring, the pathogen can move systemically from the leaf to the young stem via the leaf petiole (Serizawa and Ichikawa, 1993c). The systemic displacement of the bacterium within the young twig most probably occurs via the xylem vessels (Spinelli et al., 2011), and this represents an uncommon route of plant colonization for a P. syringae pathovar, although systemic migration via the xylem has also been described for P. s. pv. syringae in Prunus salicina shoots (Roos and Hattingh, 1987). In the summer, with temperatures increasing above 25 °C, the degree of infection is drastically reduced, even though the bacterium can colonize the host through the stomata and hydathodes (Serizawa and Ichikawa, 1993b, 1993c). During the autumn, the lenticels and buds are the main colonization sites for the pathogen (Serizawa and Ichikawa, 1994).
Repeated isolations performed over 2 years in central Italy from green‐fleshed and yellow‐fleshed kiwifruit orchards affected by bacterial canker largely confirmed that the observations carried out in Japan are also valid for the areas characterized by a typical Mediterranean climate. However, in the case of heavy infection, canker formation along the leaders and trunk can also occur during the summer. Moreover, the bacterium can colonize the fruit stalk after harvest. Remarkably, field surveys carried out in Italy in recent years have clearly indicated that some agronomical techniques that cause wounds can greatly favour the penetration of the pathogen within the plant. Indeed, the binding of young twigs to poles, the irrigation tube touching the shoots and pruning can all facilitate bacterial colonization. In addition, winter and spring frosts and hail storms have been shown to greatly enhance the ability of the pathogen to spread within and between orchards. Excessive nitrogen fertilization can also induce more susceptibility in the plant to the disease. The cycle of disease of P. s. pv. actinidiae in Mediterranean areas, shown in Fig. 4, is probably different from that in New Zealand, where deep frosts are rare, the rains are more regularly distributed throughout the year and, during the summer, the temperatures are lower than those generally recorded in a Mediterranean climate.
Figure 4.
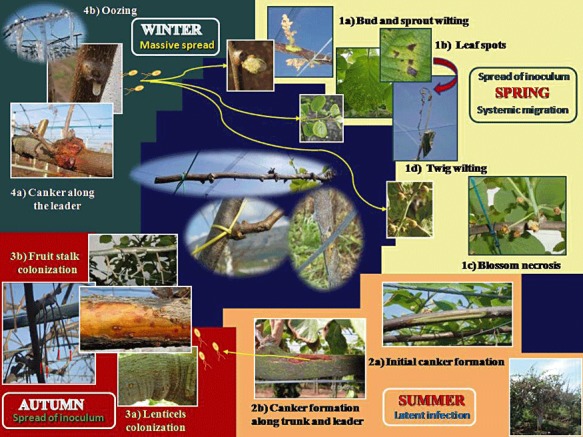
Cycle of disease of Pseudomonas syringae pv. actinidiae on Actinidia deliciosa and A. chinenis, as observed in Italy in areas characterized by a Mediterranean climate. In the middle of the panel, some agronomical techniques are indicated (i.e. winter pruning, tying of twigs, irrigation tube scraping the main young trunk) which dramatically enhance the possibility of plant colonization of the pathogen through the wounds. Frost is retained as a fundamental predisposing factor.
Infected pollen has been implicated as a source of pathogen introduction in New Zealand. Recently, the first evidence that P. s. pv. actinidiae can survive in Actinidia pollen grains has been published (Vanneste et al., 2011b) and, subsequently, confirmed (Gallelli et al., 2011b; Stefani and Giovanardi, 2011). However, it is not known how the infected pollen grains can contribute to the spread of the pathogen in the field. Noteworthy, Pseudomonas syringae pv. actinidiae shows the capability to survive in detached organs, such as leaf litter and twigs, until 45 days after their fall (Marcelletti et al., 2011; http://www.kvh.org.nz).
The putative epiphytic phase of the pathogen has yet to be fully ascertained, as some investigations suggest that the pathogen might survive epiphytically on asymptomatic flowers and leaves (Stefani and Giovanardi, 2011; Vanneste et al., 2011c), whereas others point out the failure to recover the pathogen from the leaf surface during summer (G. M. Balestra, personal communication, University of Tuscia, Viterbo, Italy). The assessment of the epiphytic and endophytic presence of the bacterium in fruits would seem to indicate a negligible risk of pathogen introduction through apparently healthy fruits. Indeed, some studies totally exclude such a possibility (Minardi et al., 2011), whereas others suggest that it is possible to find the bacterium within the fruits, although at very low levels of contamination, for samples collected from very infected orchards (Gallelli et al., 2011b). Similar to pollen, it remains to be verified whether an apparently healthy fruit could be conducive for starting the infection.
Field evidence from the recent epidemics of bacterial canker in Italy has indicated that the pandemic P. s. pv. actinidiae population is very aggressive to both A. chinensis and A. deliciosa, whereas the previously established population did not cause significant damage to A. deliciosa. The strain representative of the current pandemic population performed better than the strains from past outbreaks in Japan and Italy on A. chinensis leaves, although it also performed well on A. deliciosa. In contrast, P. s. pv. actinidiae strains from past outbreaks showed a remarkably higher multiplication trend in A. deliciosa than in A. chinensis. These results confirm the higher fitness of the pandemic population to different Actinidia species, and the fact that the strains from past outbreaks of bacterial canker had a more specific interaction with A. deliciosa (Marcelletti et al., 2011).
CONCLUDING REMARKS AND PERSPECTIVES
The enormous efforts currently in progress in Italy and New Zealand are geared towards gaining information that can be used to control the pandemics caused by P. s. pv. actinidiae. These efforts are focusing on an understanding of the pathogen and host genomes, on the one hand, and on the life cycle of the bacterium and field strategies to prevent and control the epidemic on the other, with the aim being to develop an effective strategy to eliminate or, at least, greatly reduce the worldwide risk of P. s. pv. actinidiae outbreaks during the cultivation of Actinidia species. By starting with studies of the sudden and unexpected pandemic of kiwifruit bacterial canker, much useful and practical information has already been obtained. In Italy, in some well‐managed orchards, coexistence with the disease by following precise, scheduled spray treatments, coupled with improved agronomical and preventive techniques, is a reality. However, taking into consideration the different climatic conditions occurring in the different areas of kiwifruit production worldwide, a single, general strategy is not sufficient for the control of this pathogen. For these reasons, studies should continue to investigate all aspects of the disease for the development of finely tuned strategies.
The development of genetic tolerance to bacterial canker in the kiwifruit (i.e. A. deliciosa and A. chinensis) germplasm, including the pollinators, seems to be very important to better face the possible endemic presence of the pathogen in all areas of cultivation. However, the achievement of this goal will require several years, as all of the current cultivated germplasm is quite susceptible to the disease. It is also clear that, in the presence of the bacterium, A. chinensis cannot be cultivated in areas characterized as having winter and/or spring frosts. A real climatic assessment for the determination of suitable areas seems necessary to continue the production of this valuable crop. The development of predictive model(s) based on climatic parameters and on the cycle of disease of the bacterium, with the aim to advise farmers of a possible pathogen infection, could also help to improve the control strategy of the disease. Strict phytosanitary measures based on national/regional laws should be applied in all areas of cultivation to effectively reduce the spread of the pathogen. Finally, the adoption on a large scale of an effective certification scheme that enables the assessment of the possible presence of P. s. pv. actinidiae in apparently healthy propagative material seems to be fundamental for a safer global circulation of kiwifruit plants.
ACKNOWLEDGEMENTS
The authors declare no conflicts of interest.
REFERENCES
- Abelleira, A. , Lopez, M.M. , Penalver, J. , Aquin, O. , Mansilla, J.P. , Picoaga, A. and Garcia, M.J. (2011) First report of bacterial canker of kiwifruit caused by Pseudomonas syringae pv. actinidiae in Spain. Plant Dis. 95, 1583. [DOI] [PubMed] [Google Scholar]
- Anderson, P.K. , Cunningham, A.A. , Patel, N.G. , Morales, F.J. , Epstein, P.R. and Daszak, P. (2004) Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 19, 535–544. [DOI] [PubMed] [Google Scholar]
- Arnold, D.L. and Jackson, R.W. (2011) Bacterial genomes: evolution of pathogenicity. Curr. Opin. Plant Biol. 14, 385–391. [DOI] [PubMed] [Google Scholar]
- Arnold, D.L. , Jackson, R.W. , Waterfield, N.R. and Mansfield, J.W. (2007) Evolution of microbial virulence: the benefits of stress. Trends Genet. 23, 293–300. [DOI] [PubMed] [Google Scholar]
- Australian Government (2011) Pest risk analysis report for Pseudomonas syringae pv. actinidiae associated with Actinidia (kiwifruit) propagative material (draft). July 2011, 41 pp.
- Balestra, G.M. , Mazzaglia, A. , Quattrucci, A. , Renzi, M. and Rossetti, A. (2009) Current status of bacterial canker spread on kiwifruit in Italy. Australas. Plant Pathol. 4, 34–36. [Google Scholar]
- Balestra, G.M. , Renzi, M. and Mazzaglia, A. (2010) First report of bacterial canker of Actinidia deliciosa caused by Pseudomonas syringae pv. actinidiae in Portugal. New Dis. Rep. 22, 10. [Google Scholar]
- Baltrus, D.A. , Nishimura, M.T. , Romanchuk, A. , Chang, J.H. , Mukhtar, M.S. , Cherkis, K. , Roach, J. , Grant, S.R. , Jones, C.D. and Dangl, J.L. (2011) Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog. 7, e1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastas, K.K. and Karakaya, A. (2012) First report of bacterial canker of kiwifruit caused by Pseudomonas syringae pv. actinidiae in Turkey. Plant Dis. 96, 452. [DOI] [PubMed] [Google Scholar]
- Brotz, H. and Sahl, H‐G. (2000) New insights into the mechanism of action of antibiotics‐diverse biological effects by binding to the same molecular target. J. Antimicrob. Chem. 46, 1–6. [DOI] [PubMed] [Google Scholar]
- Bull, C.T. , De Boer, S.H. , Denny, T.P. , Firrao, G. , Fischer‐Le Saux, M. , Saddler, G.S. , Scortichini, M. , Stead, D.E. and Takikawa, Y. (2010) Comprehensive list of names of plant pathogenic bacteria, 1980–2007. J. Plant Pathol. 92, 551–592. [Google Scholar]
- Carniel, E. (2001) The Yersinia high‐pathogenicity island: an iron‐uptake island. Microb. Infect. 3, 561–569. [DOI] [PubMed] [Google Scholar]
- Chapman, J. , Taylor, R. and Alexander, B. (2011) Second report on characterisation of Pseudomonas syringae pv. actinidiae (Psa) isolates in New Zealand. Ministry of Agriculture and Forestry. [Google Scholar]
- Cooksey, D.A. (1993) Molecular mechanism of copper resistance and accumulation in bacteria. FEMS Microbiol. Rev. 14, 381–386. [DOI] [PubMed] [Google Scholar]
- Daszak, P. , Cunningham, A.A. and Hyatt, A.D. (2000) Emerging infectious diseases of wildlife—threats to biodiversity and human health. Science, 284, 1311. [DOI] [PubMed] [Google Scholar]
- Delledonne, M. , Xia, Y. , Dixon, R.A. and Lamb, C. (1998) Nitric oxide functions as a signal in plant disease resistance. Nature, 394, 585–588. [DOI] [PubMed] [Google Scholar]
- European Plant Protection Organization (2011a) First report of Pseudomonas syringae pv. actinidiae in Chile. EPPO Reporting Service. n°3, 2011/055. Available at http://www.eppo.org. [accessed on February 14, 2012]. [Google Scholar]
- European Plant Protection Organization (2011b) First report of Pseudomonas syringae pv. actinidiae in Switzerland. EPPO Reporting Service. n°8, 2011/168. Available at http://www.eppo.org. [accessed on February 14, 2012]. [Google Scholar]
- Everett, K.R. , Taylor, R.K. , Romberg, M.K. , Rees‐George, J. , Fullerton, R.A. , Vanneste, J.L. and Manning, M.A. (2011) First report of Pseudomonas syringae pv. actinidiae causing kiwifruit bacterial canker in New Zealand. Aust. Plant Dis. Notes, 6, 67–71. [Google Scholar]
- Ferrante, P. and Scortichini, M. (2009) Identification of Pseudomonas syringae pv. actinidiae as causal agent of bacterial canker of yellow kiwifruit (Actinidia chinensis Planchon) in central Italy. J. Phytopathol. 157, 768–770. [Google Scholar]
- Ferrante, P. and Scortichini, M. (2010) Molecular and phenotypic features of Pseudomonas syringae pv. actinidiae isolated during recent epidemics of bacterial canker on yellow kiwifruit (Actinidia chinensis) in central Italy. Plant Pathol. 69, 954–962. [Google Scholar]
- Ferrante, P. and Scortichini, M. (2011) Molecular and phenotypic variability of Pseudomonas avellanae, P. syringae pv. actinidiae and P. syringae pv. theae: the genomospecies 8 sensu Gardan et al. (1999). J. Plant Pathol. 93, 659–666. [Google Scholar]
- Gallelli, A. , L'Aurora, A. and Loreti, S. (2011a) Gene sequence analysis for the molecular detection of Pseudomonas syringae pv. actinidiae: developing diagnostic protocols. J. Plant Pathol. 93, 425–435. [Google Scholar]
- Gallelli, A. , Talocci, S. , L'Aurora, A. and Loreti, S. (2011b) Detection of Pseudomonas syringae pv. actinidiae, causal agent of bacterial canker of kiwifruit, from symptomless fruits, and twigs, and from pollen. Phytopathol. Medit. 50, 473–483. [Google Scholar]
- Gardan, L. , Shafik, H. , Belouin, S. , Broch, R. , Grimont, F. and Grimont, P.A.D. (1999) DNA relatedness among the pathovars of Pseudomonas syringae and description of Pseudomonas tremae sp. nov. and Pseudomonas cannabina sp. nov. (ex Sutic and Dowson 1959). Int. J. Syst. Bacteriol. 49, 469–478. [DOI] [PubMed] [Google Scholar]
- Goto, M. , Hikota, T. , Nakajima, M. , Takikawa, Y. and Tsuyumu, S. (1994) Occurrence and properties of copper‐resistance in plant pathogenic bacteria. Ann. Phytopathol. Soc. Jpn. 60, 147–153. [Google Scholar]
- Green, S. , Studholme, D.J. , Laue, B.E. , Dorati, F. , Lovell, H. , Arnold, D. , Cottrell, J.E. , Bridgett, S. , Blaxter, M. , Huitema, E. , Thwaites, R. , Sharp, P.M. , Jackson, R.W. and Kamoun, S. (2010) Comparative genome analysis provides insights into the evolution and adaptation of Pseudomonas syringae pv. aesculi on Aesculus hippocastanum . PLoS ONE, 5, e10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, H.S. , Koh, Y.J. , Hur, J.‐S. and Jung, J.S. (2003a) Identification and characterization of coronatine‐producing Pseudomonas syringae pv. actinidiae . J. Microbiol. Biotechnol. 13, 110–118. [Google Scholar]
- Han, H.S. , Nam, H.Y. , Koh, Y.J. , Hur, J.‐S. and Jung, J.S. (2003b) Molecular bases of high‐level streptomycin resistance in Pseudomonas marginalis and Pseudomonas syringae pv. actinidiae . J. Microbiol. 41, 16–21. [Google Scholar]
- Han, H.S. , Koh, Y.J. , Hur, J.‐S. and Jung, J.S. (2004) Occurrence of the strA‐strB streptomycin resistance genes in Pseudomonas species isolated from kiwifruit plants. J. Microbiol. 42, 365–368. [PubMed] [Google Scholar]
- Jackson, R.W. , Athanassopoulos, E. , Tsiamis, G. , Mansfield, J.W. , Sesma, A. , Arnold, D.L. , Gibbon, M.J. , Murillo, J. , Taylor, J.D. and Vivian, A. (1999) Identification of a pathogenicity island, which contains genes for virulence and avirulence, on a large native plasmid in the bean pathogen Pseudomonas syringae pathovar phaseolicola . Proc. Natl. Acad. Sci. USA, 96, 10 875–10 880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, R.W. , Johnson, L.J. , Clarke, S.R. and Arnold, D.L. (2011a) Bacterial pathogen evolution: breaking news. Trends Genet. 27, 32–40. [DOI] [PubMed] [Google Scholar]
- Jackson, R.W. , Vinatzer, B. , Arnold, D.L. , Dorus, S. and Murillo, J. (2011b) The influence of the accessory genome on bacterial pathogen evolution. Mobile Genet. Elem. 1, 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh, J.K. and Nou, I. (2002) DNA markers for identification of Pseudomonas syringae pv. actinidiae . Mol. Cells, 13, 309–314. [PubMed] [Google Scholar]
- Koh, J.K. , Cha, B.J. , Chung, H.J. and Lee, D.H. (1994) Outbreak and spread of bacterial canker in kiwifruit. Korean J. Plant Pathol. 10, 68–72. [Google Scholar]
- Koh, Y.J. , Kim, G.H. , Jung, J.S. , Lee, Y.S. and Hur, J.S. (2010) Outbreak of bacterial canker on Hort16A (Actinidia chinensis Planchon) caused by Pseudomonas syringae pv. actinidiae in Korea. N. Z. J. Crop Hort. Sci. 38, 275–282. [Google Scholar]
- Lee, J.H. , Kim, J.H. , Kim, G.H. , Jung, J.S. , Hur, J.‐S. and Koh, Y.J. (2005) Comparative analysis of Korean and Japanese strains of Pseudomonas syringae pv. actinidiae causing bacterial canker of kiwifruit. Plant Pathol. J. 21, 119–126. [Google Scholar]
- Liang, Y. , Zhang, X. , Tian, C. , Gao, A. and Wang, P. (2000) Pathogenic identification of kiwifruit bacterial canker in Shaanxi. J. Northwest Forestry College, unpaginated. Available at: http://en.cnki.com.cn/Article_en/CJFDTOTAL‐XBLX200001006.htm[accessed on February 14, 2012].
- Manceau, C. and Brin, C. (2003) Pathovars of Pseudomonas syringae are structured in genetic populations allowing the selection of specific markers for their detection in plant samples In: Pseudomonas Syringae and Related Pathogens (Iacobellis N.S., Collmer A., Hutcheson S.W., Mansfield J.W., Morris C.E., Murillo J., Schaad N.W., Stead D.E., Surico G. and Ulrich M.S., eds), pp. 503–512. Dordrecht: Kluwer Academic Publishers. [Google Scholar]
- Marcelletti, S. and Scortichini, M. (2011) Clonal outbreaks of bacterial canker caused by Pseudomonas syringae pv. actinidiae on Actinidia chinensis and A. deliciosa in Italy. J. Plant Pathol. 93, 479–483. [Google Scholar]
- Marcelletti, S. , Ferrante, P. , Petriccione, M. , Firrao, G. and Scortichini, M. (2011) Pseudomonas syringae pv. actinidiae draft genome comparisons reveal strain‐specific features involved in adaptation and virulence to Actinidia species. PLoS ONE, 6, e27297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minardi, P. , Lucchese, C. , Ardizzi, S. and Mazzucchi, U. (2011) Evidence against the presence of Pseudomonas syringae pv. actinidiae in fruits of Actinidia orchards affected by bacterial canker. J. Plant Pathol. 93 (Supplement 4), 43. [Google Scholar]
- Nakajima, M. , Yamashita, S. , Takikawa, Y. , Tsuyumu, S. , Hibi, T. and Goto, M. (1995) Similarity of streptomycin resistance gene(s) in Pseudomonas syringae pv. actinidiae with strA and strB of plasmid RSF1010. Ann. Phytopathol. Soc. Jpn. 61, 489–492. [Google Scholar]
- Nakajima, M. , Goto, M. and Hibi, T. (2002) Similarity between copper resistance genes from Pseudomonas syringae pv. actinidiae and P. syringae pv. tomato . J. Gen. Plant Pathol. 68, 68–74. [Google Scholar]
- Neilands, J.B. (1995) Siderophores—structure and functions of microbial iron transport compounds. J. Biol. Chem. 270, 26 723–26 726. [DOI] [PubMed] [Google Scholar]
- Raymond, K.N. , Dertz, E.A. and Kim, S.S. (2003) Enterobactin: an archetype for microbial iron transport. Proc. Natl. Acad. Sci. USA, 100, 3584–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees‐George, J. , Vanneste, J.L. , Cornish, D.A. , Pushparajah, I.P.S. , Yu, J. , Templeton, M.D. and Everett, K.R. (2010) Detection of Pseudomonas syringae pv. actinidiae using polymerase chain reaction (PCR) primers based on the 16S–23S rDNA intertranscribed spacer region and comparison with PCR primers based on other gene regions. Plant Pathol. 49, 453–464. [Google Scholar]
- Rodriguez‐Palenzuela, P. , Matas, I.M. , Murillo, J. , Lopez‐Solanilla, E. , Bardaji, L. , Pérez‐Martinez, I. , Rodriguez‐Moskera, M.E. , Penyalver, R. , Lopez, M.M. , Quesada, J.M. , Biehl, B.S. , Perna, N.T. , Glasner, J.D. , Cabot, E.L. , Neeno‐Eckwall, E. and Ramos, C. (2010) Annotation and overview of the Pseudomonas savastanoi pv. savastanoi NCPPB 3335 draft genome reveals the virulence gene complement of a tumour‐inducing pathogen of woody host. Environ. Microbiol. 12, 1604–1620. [DOI] [PubMed] [Google Scholar]
- Ronning, C.M. , Losada, L. , Brinkac, L. , Inman, J. , Ulrich, R.L. , Schell, M. , Nierman, W.C. and De Shazer, D. (2010) Genetic and phenotypic diversity in Burkholderia: contributions by prophage and phage‐like elements. BMC Microbiol. 10, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos, I.M.M. and Hattingh, M.J. (1987) Systemic invasion of plum leaves and shoots by Pseudomonas syringae pv. syringae introduced into petioles. Phytopathology, 77, 1253–1257. [Google Scholar]
- Sawada, H. , Takeuchi, T. and Matsuda, I. (1997) Comparative analysis of Pseudomonas syringae pv. actinidiae and pv. phaseolicola based on phaseolotoxin‐resistant ornithine carbamoyltransferase gene (argK) and 16S‐23S rRNA intergenic spacer sequences. Appl. Environ. Microbiol. 63, 282–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada, H. , Suzuki, F. , Matsuda, I. and Saitou, N. (1999) Phylogenetic analysis of Pseudomonas syringae pathovars suggests the horizontal gene transfer of argK and the evolutionary stability of hrp gene cluster. J. Mol. Evol. 49, 627–644. [DOI] [PubMed] [Google Scholar]
- Scortichini, M. (1994) Occurrence of Pseudomonas syringae pv. actinidiae on kiwifruit in Italy. Plant Pathol. 43, 1035–1038. [Google Scholar]
- Scortichini, M. , Marchesi, U. and Di Prospero, P. (2002) Genetic relatedness among Pseudomonas avellanae, P. syringae pv. theae and P. s. pv. actinidiae, and their identification. Eur. J. Plant Pathol. 108, 269–278. [Google Scholar]
- Serizawa, S. and Ichikawa, T. (1993a) Epidemiology of bacterial canker of kiwifruit. 3. The seasonal changes of bacterial population in lesions and of its exudation from lesions. Ann. Phytopathol. Soc. Jpn. 59, 469–476. [Google Scholar]
- Serizawa, S. and Ichikawa, T. (1993b) Epidemiology of bacterial canker of kiwifruit. 2. The most suitable times and environments for infection on new canes. Ann. Phytopathol. Soc. Jpn. 59, 460–468. [Google Scholar]
- Serizawa, S. and Ichikawa, T. (1993c) Epidemiology of bacterial canker of kiwifruit. 1. Infection and bacterial movement in tissue of new canes. Ann. Phytopathol. Soc. Jpn. 59, 452–459. [Google Scholar]
- Serizawa, S. and Ichikawa, T. (1994) Epidemiology of bacterial canker of kiwifruit. 5. Effect of infection in fall to early winter on the disease development in branches and trunk after winter. Ann. Phytopathol. Soc. Jpn. 60, 237–244. [Google Scholar]
- Serizawa, S. , Ichikawa, T. , Takikawa, Y. , Tsuyumu, S. and Goto, M. (1989) Occurrence of bacterial canker of kiwifruit in Japan: description of symptoms, isolation of the pathogen and screening of bactericides. Ann. Phytopatol. Soc. Jpn. 55, 427–436. [Google Scholar]
- Spinelli, F. , Donati, I. , Vanneste, J.L. , Costa, M. and Costa, G. (2011) Real time monitoring of the interactions between Pseudomonas syringae pv. actinidiae and Actinidia species. Acta Hortic. 913, 461–465. [Google Scholar]
- Stefani, E. and Giovanardi, D. (2011) Dissemination of Pseudomonas syringae pv. actinidiae through pollen and its epiphytic life on leaves and fruits. Phytopathol. Medit. 50, 501–505. [Google Scholar]
- Sundin, G.W. (2007) Genomic insights into the contribution of phytopathogenic bacterial plasmids to the evolutionary history of their hosts. Annu. Rev. Phytopathol. 45, 129–151. [DOI] [PubMed] [Google Scholar]
- Takikawa, Y. , Serizawa, S. , Ichikawa, T. , Tsuyumu, S. and Goto, M. (1989) Pseudomonas syringae pv. actinidiae pv. nov.: the causal bacterium of canker of kiwifruit in Japan. Ann. Phytopathol. Soc. Jpn. 55, 437–444. [Google Scholar]
- Ushiyama, K. , Suyama, K. , Kita, N. , Aono, N. and Fujii, H. (1992a) Isolation of kiwifruit canker pathogen, Pseudomonas syringae pv. actinidiae from leaf spot of tara vine (Actinidia arguta Planch.). Ann. Phytopathol. Soc. Jpn. 58, 476–479. [Google Scholar]
- Ushiyama, K. , Kita, N. , Suyama, K. , Aono, N. , Ogawa, J. and Fujii, H. (1992b) Bacterial canker disease of wild Actinidia plants as the infection source of outbreak of bacterial canker of kiwifruit caused by Pseudomonas syringae pv. actinidiae . Ann. Phytopathol. Soc. Jpn. 58, 426–430. [Google Scholar]
- Van Sluys, M.A. , De Oliveira, M.C. , Monteiro‐Vitorello, C.B. , Miyaki, C.Y. , Furlan, R.L. , Camargo, L.E.A. , da Silva, A.C.R. , Moon, D.H. , Takita, M.A. , Lemos, E.G.M. , Machado, M.A. , Ferro, M.I.T. , da Silva, F.R. , Goldman, M.H.S. , Goldman, G.H. , Lemos, M.V.F. , El‐Dorry, H. , Tsai, S.M. , Carrer, H. , Carraro, D.M. , de Oliveira, R.C. , Nunes, L.R. , Siqueira, W.H. , Coutinho, L.L. , Kimura, E.T. , Ferro, E.S. , Harakawa, R. , Kuramae, E.E. , Marino, C.L. , Giglioti, E. , Abreu, I.L. , Alves, L.M.C. , do Amaral, A.M. , Baia, G.S. , Blanco, S.R. , Brito, M.S. , Cannavan, F.S. , Celestino, A.V. , da Cuhna, A.F. , Fenille, R.C. , Ferro, J.A. , Formighieri, E.F. , Kishi, L.T. , Leoni, S.G. , Oliveira, A.R. , Rosa jr., V.E. , Sassaki, F.T. , Sena, J.A.D. , de Souza, A.A. , Truffi, D. , Tsukumu, F. , Yanai, G.M. , Zaros, L.G. , Civerolo, E.L. , Simpson, A.J.G. , Almeida jr., N.F. , Setubal, J.C. and Kitajima, J.P. (2003) Comparative analysis of the complete genome sequences of Pierce's disease and citrus variegated chlorosis strains of Xylella fastidiosa . J. Bacteriol. 185, 1018–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste, J.L. , Poliakoff, F. , Audusseau, C. , Cornish, D.A. , Paillard, S. , Rivoal, C. and Yu, J. (2011a) First report of Pseudomonas syringae pv. actinidiae, the causal agent of bacterial canker of kiwifruit in France. Plant Dis. 95, 1311. [DOI] [PubMed] [Google Scholar]
- Vanneste, J.L. , Giovanardi, D. , Yu, J. , Cornish, D.A. , Kay, C. , Spinelli, F. and Stefani, E. (2011b) Detection of Pseudomonas syringae pv. actinidiae in pollen samples. N. Z. Plant Protect. 64, 246–251. [Google Scholar]
- Vanneste, J.L. , Yu, J. , Cornish, D.A. , Max, S. and Clark, G. (2011c) Presence of Pseudomonas syringae pv. actinidiae, the causal agent of bacterial canker of kiwifruit on symptomatic and asymptomatic tissues of kiwifruit. N. Z. Plant Protect. 64, 241–245. [Google Scholar]
- Wang, Z. , Tang, X. and Liu, S. (1992) Identification of the pathogenic bacterium for bacterial canker on Actinidia in Sichuan. J. Southwest Agricultural University, unpaginated. Available at: http://en.cnki.com.cn/Article_en/CJFDTOTAL‐XNND199206007.htm. [accessed on February 14, 2012].


