Fusarium Vascular Wilts Caused by Fusarium Oxysporum Pose Serious Threats to Global Food Security
An adequate supply of healthy plants is essential to feed a growing population. Many plant pathogens are highly destructive, and consequently threaten global food security. The negative impact of plant pathogens on human society was illustrated by the Irish potato famine of the 1840s, the Bengal famine of the 1940s and the US Southern corn leaf blight epidemic of 1970–1971. These examples, recorded in history accounts and textbooks, may seem remote to us; however, the devastation caused by plant pathogens is not limited to the past.
Today, plant pathogens continue to hamper agricultural production and threaten the wellbeing of human society. One such example is the recent spread of Panama disease of banana, caused by Fusarium oxysporum f. sp. cubense (Foc) tropic race 4 (TR4), from South‐East Asia to Mozambique and Jordan (Butler, 2013). The first Panama disease outbreak caused by Foc race 1 almost destroyed the banana industry in the 1950s. The crisis was resolved by replacing the susceptible ‘Gros Michel’ cultivar with the race 1‐resistant Cavendish cultivar (Ploetz, 2006). Unfortunately, the newly evolved TR4 is highly virulent to Cavendish. As bananas and plantains are significant staple foods and represent a primary dietary source of carbohydrates in Africa, South‐East Asia and tropical America, the spread of Panama disease could potentially create localized food shortages, intensify world hunger and exacerbate poverty in developing nations.
The causal agent of Panama disease, Foc, is one member of the root‐infecting fungal pathogens in the F. oxysporum species complex (FOSC). Members within this species complex cause destructive and intractable Fusarium vascular wilts in over 100 plant species, ranging from gymnosperms to angiosperms and monocots to dicots. Listed as one of the top 10 fungal pathogens, diseases caused by F. oxysporum challenge the production of numerous economically important crops, including banana (Musa acuminata), cotton (Gossypium hirsutum), canola (Brassica napus) and tomato (Solanum lycopersicum) (Ma et al., 2013).
As a species complex, F. oxysporum has a very broad host range. Interestingly, individual F. oxysporum isolates often exhibit a high degree of host specificity; a single pathogenic form usually infects a single plant species. Isolates with pathogenicity for the same host are grouped into the same forma specialis. For instance, isolates that are pathogenic to banana plants are classified as F. oxysporum f. sp. cubense (Foc) and those that infect tomato plants are classified as F. oxysporum f. sp. lycopersici (Fol).
Horizontally Acquired Chromosomes Define Host‐Specific Pathogenicity in the FOSC
An insightful comparative study using one F. oxysporum strain isolated from a diseased tomato plant revealed structural and functional compartmentalization of the genome (Ma et al., 2010) (Fig. 1). This finding was further confirmed by optical mapping and genomic sequencing of all F. oxysporum strains examined to date, including a TR4 strain isolated from the recent outbreak of Panama disease (L.‐J. Ma, unpublished data).
Figure 1.
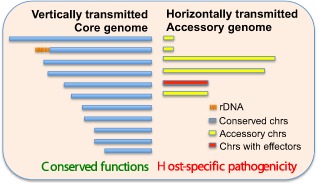
Structural and functional compartmentalization of the Fusarium oxysporum f. sp. lycopersici genome. The genome is divided into: (1) the ‘core’ genome, which is vertically transmitted, performs conserved functions and is almost identical among all F. oxysporum strains; and (2) the ‘accessory’ genome, which is horizontally transmitted, responsible for host‐specific pathogenicity and unique to each forma specialis. chr, chromosome.
Structurally, the F. oxysporum genome can be divided into the ‘core’ and the ‘accessory’ genomic regions. The ‘core’ genomic regions, which reside on conserved chromosomes, are preserved among all Fusarium species and are present in all F. oxysporum strains, regardless of their hosts. The conservation of the ‘core’ indicates vertical transmission. In contrast, the ‘accessory’ genomic regions, in the form of transposon‐rich pathogenicity chromosomes, are absent from other sequenced Fusarium genomes, except those that share the same host. These pathogenicity chromosomes are horizontally transferred and contribute directly to the host specificity.
The structural compartmentalization supports the functional partition of the genome, as the vertically transmitted ‘core’ genomic regions perform all of the essential functions and the horizontally transferred ‘accessory’ genomic regions encode host‐specific virulent factors (such as effectors), but lack housekeeping genes involved in primary metabolism. The experimental transfer of entire chromosomes introduces pathogenicity to a genetically distinct, non‐pathogenic isolate (Ma et al., 2010), strengthening the conclusion that horizontally transferred chromosomes determine host‐specific pathogenicity in the FOSC.
The mechanism of horizontal transfer also explains the polyphyletic origins of host specificity in the FOSC and the rapid emergence of new pathogenic lineages in distinct genetic backgrounds. However, this atypical mechanism introduces genetic variation rapidly and at a large scale. As the introduction of virulence factors through horizontal chromosome transfer is contrary to traditional origins of genetic diversity in eukaryotes, there is a need to develop novel strategies to control and manage this group of notorious pathogens.
Host‐Specific Virulence Factors Encoded in the ‘Accessory’ Genomic Regions Provide Novel Means to Diagnose Fusarium Vascular Wilts
The proactive prevention of the introduction of a disease agent (pathogen) into a region in which it does not occur is the target of all disease management programmes. This is particularly true for the control of Fusarium vascular wilts, as the elimination of this group of pathogens is almost impossible once they have been introduced (see next section). Effective prevention depends on an accurate diagnosis and precise forecasting of the potential pathogen threats. Because genes encoded in the horizontally transferred chromosomes control the host‐specific pathogenicity of the FOSC, conventional diagnostic approaches, either based on morphological characterization or through phylogenetic affiliation, may not always produce dependable results.
The fact that host‐specific virulence factors, such as effectors, are encoded in the ‘accessory’ region of the genome enables the use of host‐specific effectors to identify specific pathotypes within the FOSC. The method was developed to identify the tomato pathogen Fol and to discriminate this pathogen from other formae speciales based on the understanding of gene‐for‐gene interactions between Fol and tomato (Lievens et al., 2009). Given the structural and functional compartmentalization of the F. oxysporum genome, quests for host‐specific effectors should focus on small ‘accessory’ genomic regions. If chromosomal mapping information is unavailable, the ‘accessory’ regions can be broadly defined by removing large portions of the genome assembly that belong to the ‘core’. The elimination of genomic regions may sound like a minor detail in data processing. However, this simple procedure narrows the search for host‐specific effectors to specific regions with high confidence, thereby simplifying the search for host‐specific effectors from what may be likened to a treasure hunt in unexplored territory to a road trip with a coarsely defined road map.
Enhancement of Plant Defence Is the Ultimate Goal in Combating Yield Loss Caused by Fusarium Vascular Wilts
Once pathogens are introduced into a region, disease control involves either the removal of pathogenic agents or the protection of plants from the damage caused by these pathogens. Members of the FOSC produce chlamydospores that can survive in soil for over 20 years. Even during the vegetative growth phase, F. oxysporum isolates usually have a high level of resistance to most fungicides, including sterol biosynthesis‐inhibiting and demethylation‐inhibiting (DMI) fungicides. Therefore, the removal of agents that cause Fusarium vascular wilts is challenging, if not impossible.
The most cost‐effective and environmentally friendly option is to develop wilt‐resistant plants. Over the years, our understanding of plant immunity, which uses two basic defence strategies (Jones and Dangl, 2006), has improved. First, plants use transmembrane pattern recognition receptors (PRRs) to recognize conserved, pathogen‐associated molecular patterns (PAMPs) and induce PAMP‐triggered immunity (PTI). Successful pathogens have evolved virulence proteins (such as small secreted effectors) to suppress PTI, and thereby cause effector‐triggered susceptibility (ETS). Second, plants recognize effectors and induce effector‐triggered immunity (ETI).
As a hemibiotrophic pathogen, F. oxysporum goes through multiple physiological transitions throughout the infection course: the fungus switches from being a saprobe (free‐living state), to a biotroph (deriving nutrients from living plant tissues), to a necrotroph (obtaining nutrients after killing the plant host). Regardless of their hosts, all F. oxysporum infections are divided into primary and secondary determinative phases. The primary determinative phase, which involves the transition from saprophytic to biotrophic growth, includes the penetration of the fungus through the epidermis into the root and colonization of the cortex and endodermis. The secondary determinative phase entails the colonization of the vascular bundles, representing the necrotrophic growth of the pathogen. Most studies of F. oxysporum–host plant interactions have addressed pathogenicity, and therefore the secondary determinative phase. If the interaction between the fungus and plant is likened to a war, the decisive battles occur during the primary determinative phase. Arguably, studies that focus on the primary determinative phase are thus more critical for understanding plant resistance.
In almost all F. oxysporum pathosystems, the primary determinative phase occurs during the first couple of days after the pathogen–host encounter. For instance, in the F. oxysporum–Arabidopsis pathosystem, the fungus penetrates the root surface in the first day post‐inoculation. Within this time window, no substantial differences between compatible (pathogenic) and incompatible (non‐pathogenic) interactions are apparent. Electron microscopy analyses revealed that plant root cells, particularly those in the superficial layers, such as the epidermis, react to fungal growth even before fungal penetration (Rodriguezgalvez and Mendgen, 1995), indicating that signals are exchanged during the initial stages of the interaction.
Fusarium oxysporum is a metropolitan fungus. Both the pathogenic and non‐pathogenic strains are widely distributed. As a result of their high levels of host specificity, most isolates are non‐pathogenic towards non‐hosts. Interestingly, all strains tested in our laboratory and non‐pathogenic mutants tested in other studies are able to penetrate the plant root surface, suggesting that initial fungal invasion has no specificity and that plants are quite effective at defending themselves against the majority of F. oxysporum strains after this initial invasion. Plant vulnerability towards a small fraction of pathogenic strains is the result of ETS, which develops when pathogens have evolved host‐specific effectors that overcome plant PTI. As the effectors produced by a pathogen can be recognized by the secondary response of its host and induce ETI, PTI and/or ETI may be involved in incompatible/resistant interactions.
As F. oxysporum isolates have almost identical core genomes, it is conceivable that different pathotypes may share the same PAMP molecules. Therefore, the identification of PRRs that recognize F. oxysporum PAMP molecules from model plants such as Arabidopsis thaliana, followed by their introduction into crop plants that lack this recognition capability, may induce broad resistance. This concept was tested by introducing an Arabidopsis receptor protein that recognizes the bacterial PAMP signal into tomato and tobacco plants. The successful transformation of the gene preserved the PAMP recognition properties of the encoded receptor protein and induced resistance to multiple bacterial pathogens in the heterologous hosts (Lacombe et al., 2010). This argues for the importance of examining the plant–fungal relationship using model plant systems to identify PAMP molecules present in the F. oxysporum species and of identifying plant proteins involved in the perception of F. oxysporum‐specific PAMPs. Severe wilt disease is the result of ETS, as fungal effectors target plant susceptibility genes to circumvent host PTI. Bioengineering either to disarm pathogens by disrupting virulence factors or to remove the plant proteins that serve as targets of fungal effectors may also lead to the development of broad‐spectrum resistant crops. Overall, an understanding of the genome dynamics that underlie pathogen adaptation should provide guiding principles for the development of management strategies.
Conclusive Remarks and Future Research
The field of genomics has revolutionized many aspects of biological research, as exemplified by the rapid developments in personalized medicine. Agricultural genomics, considered as the next wave in agricultural research after the green revolution, has the potential to translate genomic information into agronomic advancement. The combination of a focused approach that dissects plant immunity against F. oxysporum using model plants and a system approach that investigates genome evolution, organism adaptation and functional dynamics of the FOSC in diverse ecological niches will influence the development of rational strategies to control Fusarium vascular wilts.
Focused approach: dissection of plant immunity against F. oxysporum using model plants
One of the most common soil‐borne fungal species, F. oxysporum has served as a long‐standing model to study root infection diseases. Comparative studies further established F. oxysporum as a testable system for the investigation of evolutionary aspects of genome dynamics and organism adaptation (Ma et al., 2010, 2013). Both F. oxysporum–Arabidopsis and F. oxysporum–tomato pathosystems have been established.
Gene‐for‐gene interactions have been best illustrated in Fol and its host plant, tomato. In this system, three plant resistance genes against Fol, i.e. I, I‐2 and I‐3, and three corresponding avirulence factors in the Fol strain, i.e. Avr1, Avr2 and Avr3, have been identified. One direct interaction has been confirmed between Avr1 and tomato I‐2 (Houterman et al., 2009). Although more genetic/genomic resources are available for the model plant Arabidopsis, much less is known about the host–pathogen interactions in the F. oxysporum–Arabidopsis pathosystem. Six quantitative resistance loci, named RESISTANCE TO F. OXYSPORUM (RFO), were detected in the Arabidopsis genome (Diener and Ausubel, 2005), and a candidate effector sharing high sequence similarity with Fol Avr1 was detected in one of the Arabidopsis‐infecting F. oxysporum strains.
Studies of Arabidopsis–pathogen interactions have led to many breakthroughs in our understanding of plant immunity. However, many of these studies, particularly those related to fungal–plant interactions, have concentrated on fungal pathogens that infect foliar tissues. Genes and pathways engaged in pathogen recognition and host responses differ significantly between root and foliar tissues. It is critically important to continue to develop the F. oxysporum–Arabidopsis pathosystem to dissect the plant response to root‐specific infections and to understand root‐infecting pathogenesis. Studies based on model plants may have transformative effects on our ability to develop durable plants that are resistant to devastating Fusarium wilt diseases, such as Panama disease of banana.
Systems approach: investigation of ecological adaptation using systems biology
As a gene never functions in isolation, it is imperative, and now possible, to decipher its molecular function within the context of its genome, and ultimately within the complex environment in which the organism functions. Compared with investigations that focus on one or a few genes (a more traditional, reductionist approach), systems biology focuses on complex interactions within biological systems (a more holistic perspective).
One of the many strategies used in systems biology is to reconstruct the regulatory networks of an organism or of an interactive biological system. Network reconstruction of the F. oxysporum genome is particularly interesting. As this genome has undergone large‐scale horizontal gene transfer (i.e. the transfer of chromosomes), it harbours two independent, yet integrated, regulatory networks. The functional network vertically inherited from its ancestor is expected to control the essential functions of the organism. The most economical approach of controlling all genes acquired through chromosomal transfer is to regulate them through an independent regulatory network. The genetic mechanisms that integrate these two networks ultimately control the functionality of the organism, or the establishment of pathogenicity. Therefore, the understanding of such mechanisms will provide insights into network rewiring and facilitate the development of novel disease management strategies. This argument is supported by the finding that horizontally transferred genes include over 100 transcription factors, and transcription factors encoded in the conserved genomic regions (Sge1) have proven to be essential for pathogenicity and required for the expression of effector genes.
To better understand pathogen–host interactions, a systems approach may go beyond an individual genome, as regulatory network rewiring probably occurs in both the host and the pathogen in response to the flurry of signals exchanged between these two parties. A systematic dissection at the regulatory network level will highlight the key components/biological pathways participating in such interactions. One approach would be to perturb the network structure with both compatible and incompatible interactions, and then to identify shared and distinct host–pathogen protein interaction profiles for these two interactions. Such an approach would reveal how these interactions exploit distinctive or common strategies to subvert cellular pathways towards disease progression.
Furthermore, organisms do not generally live in isolation; instead, they exist in an environment rich in microbiota. One future research direction would be to systematically survey the biodiversity and structure of microbial communities within complex ecosystems related to Fusarium vascular wilts. Soil environments, in which root‐infecting wilt pathogens exist, are conducive to horizontal gene transfer. Our study demonstrated the transfer of chromosomes between isolates of F. oxysporum (Ma et al., 2010); however, the mechanisms underlying the transfer of chromosomes within and possibly among species remain to be determined. The combination of metagenomics and comparative genomics may eventually shed light on the source of these chromosomes that serve as a reservoir of all the virulence factors, and the mechanisms that mediate the transfer. An understanding of these molecular mechanisms and of the genes that control them in both the fungus and plant may facilitate the reconstruction of a balanced and beneficial microbial community in the rhizosphere.
Collectively, basic research will ultimately reveal the molecular mechanisms underlying vascular wilt diseases. The fundamental information discovered through basic research will energize translational research for the development of new and effective approaches. Considering the intractability of various Fusarium wilts and their devastating effects on crop production, the integration of fundamental research and the applied sciences will have a direct positive impact on global food security and quality of life.
References
- Butler, D. (2013) Fungus threatens top banana. Nat. Middle East 504, 195–196. [DOI] [PubMed] [Google Scholar]
- Diener, A.C. and Ausubel, F.M. (2005) RESISTANCE TO FUSARIUM OXYSPORUM 1, a dominant Arabidopsis disease‐resistance gene, is not race specific. Genetics, 171, 305–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houterman, P.M. , Ma, L. , van Ooijen, G. , de Vroomen, M.J. , Cornelissen, B.J. , Takken, F.L. and Rep, M. (2009) The effector protein Avr2 of the xylem colonizing fungus Fusarium oxysporum activates the tomato resistance protein I‐2 intracellularly. Plant J. 58, 970–978. [DOI] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Lacombe, S. , Rougon‐Cardoso, A. , Sherwood, E. , Peeters, N. , Dahlbeck, D. , van Esse, H.P. , Smoker, M. , Rallapalli, G. , Thomma, B.P. , Staskawicz, B. , Jones, J.D. and Zipfel, C. (2010) Interfamily transfer of a plant pattern‐recognition receptor confers broad‐spectrum bacterial resistance. Nat. Biotechnol. 28, 365–369. [DOI] [PubMed] [Google Scholar]
- Lievens, B. , Houterman, P.M. and Rep, M. (2009) Effector gene screening allows unambiguous identification of Fusarium oxysporum f. sp. lycopersici races and discrimination from other formae speciales. FEMS Microbiol. Lett. 300, 201–215. [DOI] [PubMed] [Google Scholar]
- Ma, L.J. , van der Does, H.C. , Borkovich, K.A. , Coleman, J.J. , Daboussi, M.J. , Di Pietro, A. , Dufresne, M. , Freitag, M. , Grabherr, M. , Henrissat, B. , Houterman, P.M. , Kang, S. , Shim, W.B. , Woloshuk, C. , Xie, X. , Xu, J.R. , Antoniw, J. , Baker, S.E. , Bluhm, B.H. , Breakspear, A. , Brown, D.W. , Butchko, R.A. , Chapman, S. , Coulson, R. , Coutinho, P.M. , Danchin, E.G. , Diener, A. , Gale, L.R. , Gardiner, D.M. , Goff, S. , Hammond‐Kosack, K.E. , Hilburn, K. , Hua‐Van, A. , Jonkers, W. , Kazan, K. , Kodira, C.D. , Koehrsen, M. , Kumar, L. , Lee, Y.H. , Li, L. , Manners, J.M. , Miranda‐Saavedra, D. , Mukherjee, M. , Park, G. , Park, J. , Park, S.Y. , Proctor, R.H. , Regev, A. , Ruiz‐Roldan, M.C. , Sain, D. , Sakthikumar, S. , Sykes, S. , Schwartz, D.C. , Turgeon, B.G. , Wapinski, I. , Yoder, O. , Young, S. , Zeng, Q. , Zhou, S. , Galagan, J. , Cuomo, C.A. , Kistler, H.C. and Rep, M. (2010) Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature, 464, 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L.‐J. , Geiser, D.M. , Proctor, R.H. , Rooney, A.P. , O'Donnell, K. , Trail, F. , Gardiner, D.M. , Manners, J.M. and Kazan, K. (2013) Fusarium pathogenomics. Annu. Rev. Microbiol. 67, 399–416. [DOI] [PubMed] [Google Scholar]
- Ploetz, R.C. (2006) Fusarium wilt of banana is caused by several pathogens referred to as Fusarium oxysporum f. sp. cubense . Phytopathology, 96, 653–656. [DOI] [PubMed] [Google Scholar]
- Rodriguezgalvez, E. and Mendgen, K. (1995) The infection process of Fusarium oxysporum in cotton root tips. Protoplasma, 189, 61–72. [Google Scholar]


