Figure 2.
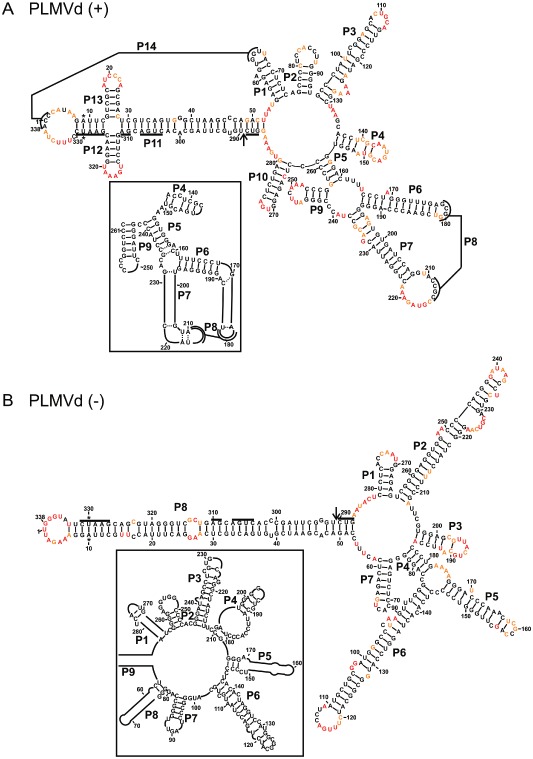
The most stable structures for both polarities of Peach latent mosaic viroid (PLMVd). The final structural models of both PLMVd (+) (A) and PLMVd (–) (B) obtained by selective 2′‐hydroxyl acylation analysed by primer extension (SHAPE) and folded by RNAstructure. The nucleotides in black denote low SHAPE reactivities (0–0.40), those in orange intermediate reactivities (0.40–0.85) and those in red highly reactive nucleotides (>0.85). The nucleotides forming the hammerhead are underlined, the cleavage site is indicated by an arrow and the mutated nucleotides are identified by stars. The insets show the differences observed when structures are compared with previously accepted models. Only regions with variations are shown in the insets. The numbering of the stems is in accordance with the first reported PLMVd structure (Bussiere et al., 2000). The numbering of the stems of the (–) polarity structure is in accordance with that used for the (+) polarity.
