Figure 1.
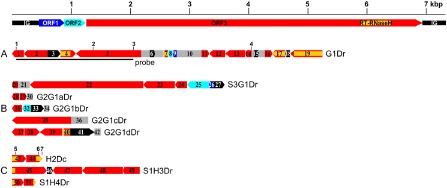
Schematic representation of badnavirus endogenous pararetroviruses (EPRVs) of Dioscorea cayenensis‐rotundata. A scaled linear view of the genome organization of Dioscorea bacilliform alata virus (DBALV) is shown in the top panel. The intergenic region (IG) and open reading frames (ORFs) appear with the following colour codes: IG, black; ORF1, dark blue; ORF2, light blue; ORF3, red. Rearranged badnavirus EPRVs of the D. cayenensis‐rotundata complex are shown in (A–C) using the same colour code. Rearranged fragments are numbered individually and the direction of the arrows indicates sequence orientation. Yellow horizontal lines and grey boxes indicate RT‐RNaseH domains of ORF3 sequences and uncharacterized sequences, respectively. Vertical numbered bars show the locations of the primers used for polymerase chain reaction (PCR) amplifications: 1, purifG1F; 2, verifG1F; 3, purifG1R; 4, verifG1R; 5, B389‐2F; 6, B389‐4R; 7, B389‐3R (see Table 1). (A) Dioscorea rotundata G1Dr sequence amplified from the GN155 plant. The part of the G1Dr sequence that was used as a probe in Southern blot experiments is shown by a horizontal bar. (B) Dioscorea rotundata sequences generated using the verifG1F/R primer pair. S3G1Dr was amplified from the Sd3 seedling and the four other sequences from the GN290 plant. (C) H2Dc sequence amplified from D. cayenensis HT1 plant, and S1H3Dr and S1H4Dr sequences amplified from D. rotundata Sd1 seedling using B389‐2F/3R and B389‐2F/4R primers, respectively.
