Figure 2.
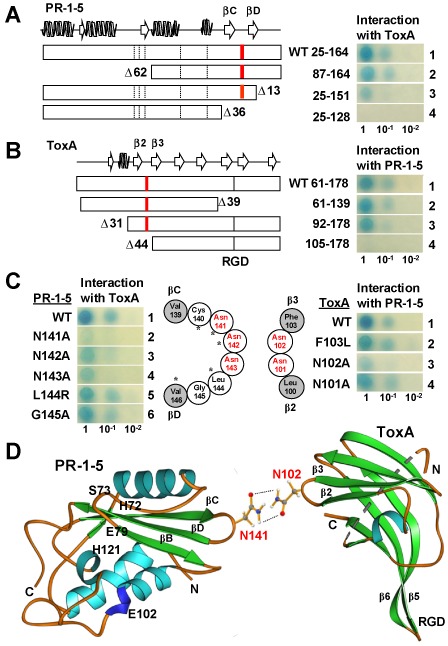
Identification of residues essential for the PR‐1‐5–ToxA interaction. (A, B) Deletion analyses. Open boxes indicate the mature proteins with secondary structures drawn at the top following P14a (Fernandez et al., 1997) and Ptr ToxA (Sarma et al., 2005) for PR‐1‐5 and SnToxA, respectively. Broken and full bars indicate the positions of the five putative active sites in the PR‐1 domain and the ToxA RGD motif, respectively. Red bars indicate the positions of the asparagine‐rich loops. Residue numbering is based on the full‐length PR‐1‐5 protein (GenBank accession number HQ541965) and SnToxA (DQ423483) proteins with ‘Δ’ indicating the deleted residues. (C) Site‐specific mutagenesis. Open and filled circles indicate residues consisting of the loops and the connecting β‐strands, respectively. Asparagine residues are highlighted in red. Asterisks indicate residues conserved in P14a. (D) Ribbon diagrams showing the predicted site‐specific interaction between PR‐1‐5 and ToxA. α helices, β strands and loops are in light blue, green and orange, respectively. The side‐chains of the two interacting asparagine residues are shown in a ‘stick–ball’ scheme with red, grey, blue and white balls for oxygen, carbon, nitrogen and hydrogen atoms, respectively. Broken lines indicate potential hydrogen bonds.
