Figure 5.
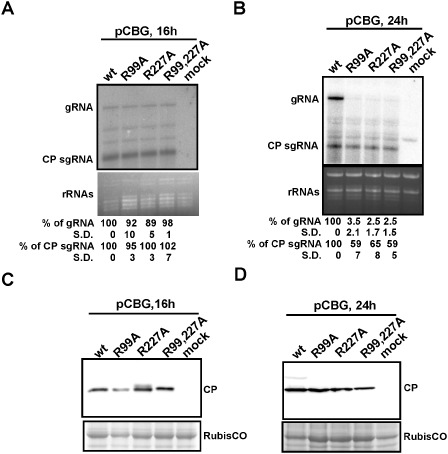
Effects of coat protein (CP) mutations on Bamboo mosaic virus (BaMV) accumulation in protoplasts. The protoplasts of Nicotiana benthamiana were inoculated with pCBG or the mutant derivatives, as indicated at the top. Total RNAs were extracted and analysed at 16 h post‐inoculation (hpi) (A) and 24 hpi (B) by Northern blot analysis using 32P‐labelled RNA probes complementary to the 3′ untranslated region (UTR) of positive‐sense BaMV RNA. Ethidium bromide‐stained rRNAs are shown as loading controls. The relative amounts (%) of viral genomic (g) and CP subgenomic (sg) RNAs, compared with that of the wild‐type (wt), are shown at the bottom. The percentage values shown represent the averages of all quantified samples. Three independent protoplast inoculation experiments were performed. The standard deviation is abbreviated as S.D. In (A), the differences between the accumulation levels of wt and each mutant were not statistically significant as analysed using Student's single‐tailed t‐test, with all P values >0.05. Total proteins were extracted from protoplasts at 16 hpi (C) and 24 hpi (D), separated by sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE), transferred to a poly(vinylidene difluoride) (PVDF) membrane and probed with antiserum specific to BaMV CP. The positions of BaMV CPs are indicated on the right. The Coomassie blue‐stained gel, showing the positions around the Ribulose‐1,5‐bisphosphate carboxylase oxygenase protein (large subunit of RubisCO), was used as a loading control.
