Figure 3.
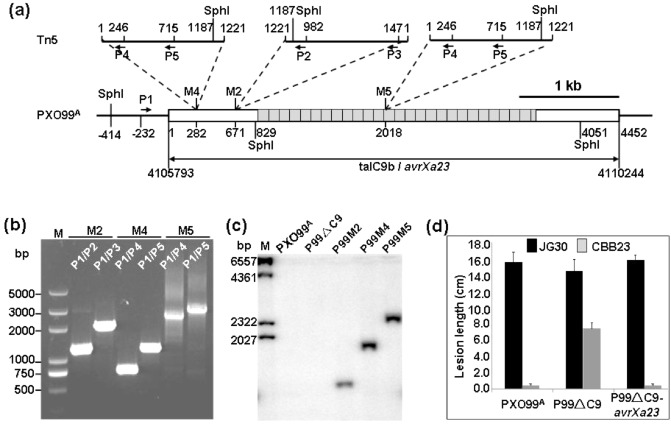
Characterization of Xanthomonas oryzae pv. oryzae (Xoo) mutants. (a) Schematic representation of insertion sites and orientations of Tn5‐DNA in mutants P99M2, P99M4 and P99M5. Black bars represent the genome of PXO99A or the Tn5‐DNA. Boxes represent talC9b located in genome region 4105793–4110244 of PXO99A (GenBank: NC_010717.1). Grey boxes represent the 26.5 repeats of talC9b. Numbers under the boxes and a nearby bar indicate the relative nucleotide positions from the first ‘A’ in the open reading frame (ORF) of talC9b. Numbers above the bars indicate the nucleotide positions of the 1221‐bp Tn5‐DNA. Broken lines indicate the Tn5 insertion sites in P99M2 (M2), P99M4 (M4) and P99M5 (M5). The nucleotide positions of the polymerase chain reaction (PCR) primers (P1–P5) and SphI restriction sites are also indicated. (b) Nested PCR analysis of mutants P99M2 (M2), P99M4 (M4) and P99M5 (M5). Primer combinations and the size (bp) of the molecular ladder (M) bands are shown. (c) Southern blotting of Xoo strains. Genomic DNA of Xoo strains were digested with SphI, separated in 1.2% (w/v) agarose gel and transferred to a nylon membrane. The 569‐bp PCR fragment amplified from Tn5‐DNA using primers P3 and P5 (a) was labelled with 32 P‐dCTP and used as the DNA probe. PXO99A and P99ΔC9 were used as negative controls. M, molecular ladder. (d) Virulence assays of the talC9b knockout mutant P99ΔC9 and its complementary strain P99ΔC9‐avrXa23 in CBB23 by artificial leaf clipping inoculation. Wild type Xoo PXO99A and susceptible rice JG30 were used as controls. The lesion length was the mean ± SD of five leaves measured at 14 days post‐inoculation.
