Figure 2.
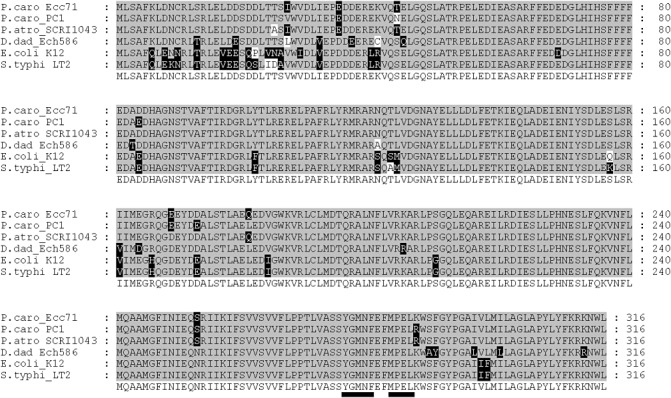
Alignment of deduced amino acid sequences of Pectobacterium carotovorum Ecc71 CorA with homologues from other bacteria. The consensus amino acid sequence from these strains is shown at the bottom of the alignment. Residues that are identical to the consensus sequence are shaded in grey, conservative substitutions are shaded in black and nonconservative substitutions are shaded in white. The signature sequences, YGMNF and MPEL, are located at the C‐terminus and are underlined in the consensus sequence. The multiple sequence alignment was generated using T‐COFEE (European Bioinformatics Institute) and the alignment output was produced using GeneDoc (Nicholas et al., 1997). Sequence IDs are abbreviated as follows: Ecc71, ‘P.caro_Ecc71’; P. carotovorum PC1(PC1 _3969), ‘P.caro_PC1’; P. atrosepticum SCRI 1043 (ECA_4177), ‘P.atro_SCRI1043’; Dicteya dadantii (Dd586_3867), ‘D.dad_Ech586’; Escherichia coli K‐12 substr. MG1655(AAA67612), ‘E.coli_K12’; Salmonella typhimurium LT2 (STM3952), ‘S.typhi_LT2’.
