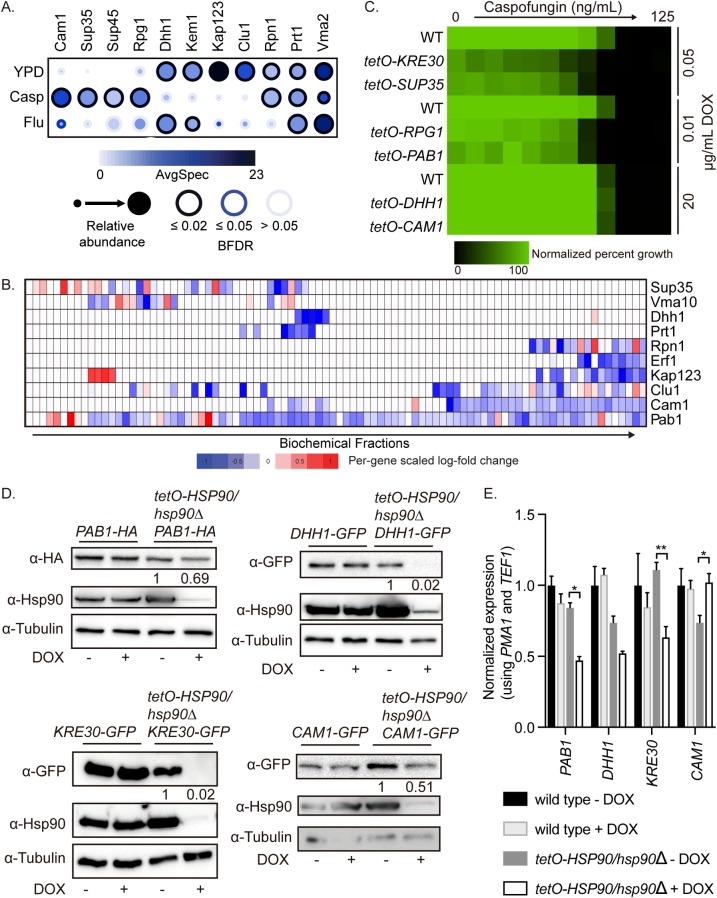
Fig 6. Hsp90 is required for the stability of stress granule and P-body proteins.
(A) Stress granule and P-body proteins vary in their interaction with Hsp90 upon drug treatment. AP-MS was performed on HSP90E36A-GFP/HSP90 cells grown at 30°C in the presence or absence of 8 μg/mL fluconazole (Flu) or 100 nM caspofungin (Casp). Statistically significant Hsp90 interaction partners are shown as a dot plot in which the node color represents the absolute spectral count, node size represents relative abundance between the conditions, and node edge color represents the SAINTexpress BFDR rate. Raw data for this figure can be found in S4 Table. (B) Stress granule protein abundance upon Hsp90 depletion. Protein abundance across 120 IEX fractions in untreated cells or cells treated with DOX to transcriptionally repress HSP90 in the tetO-HSP90/hsp90Δ strain. Raw data for this figure can be found in S3 Table. (C) Stress granule proteins are required for caspofungin tolerance. Mutants of proteins involved in stress granules (Pab1, Kre30, and Sup35) are hypersensitive to caspofungin. MIC assays were performed in YPD medium at 30°C for 24 hours, and optical densities at 600 nm were averaged for two biological replicates performed in technical duplicate. Growth was normalized to the no drug well. To repress target gene expression, the strains were incubated in the indicated concentrations of DOX. Raw data for this figure can be found in S1 Data. (D) Stress granule and P-body protein levels are decreased upon transcriptional repression of HSP90. Cells were grown overnight in ±0.5 μg/mL DOX to repress HSP90 in the tetO-HSP90/hsp90Δ strain and then subcultured into medium ± 5 μg/mL DOX for 4 hours before protein extraction and western blotting. Protein levels were normalized to the tubulin loading control and quantified compared with the no DOX control. (E) Stress granule protein transcripts are not dependent on Hsp90. Cells were grown overnight in ±0.5 μg/mL DOX to repress HSP90 in the tetO-HSP90/hsp90Δ strain and then subcultured into medium ± 5 μg/mL DOX for 4 hours before RNA extraction and qRT-PCR. Transcript levels were normalized to PMA1 and TEF1. Significance was determined by one-way ANOVA. ** indicates P value <0.01, * indicates P value <0.05. Raw data for this figure can be found in S1 Data. AP-MS, affinity purification-mass spectrometry; BFDR, Bayesian false discovery rate; DOX, doxycycline; GFP, green fluorescent protein; IEX, ion exchange; MIC, minimum inhibitory concentration; P-body, processing body; qRT-PCR, quantitative reverse transcription PCR; SAINTexpress, Significance Analysis of INTeractome; YPD, yeast extract peptone dextrose.