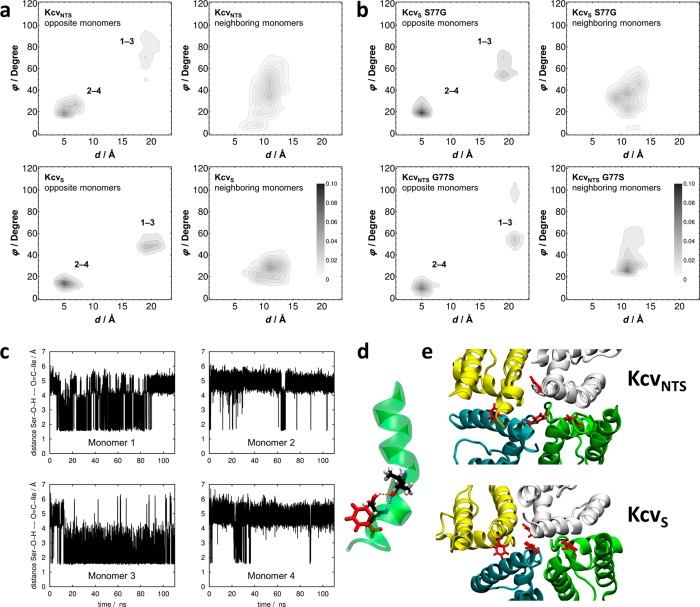
Figure 7.
Dynamics of TMD2 in KcvS/KcvNTS and mutants. (a) Distribution of distance-angle pairs characterizing π-stack geometries in KcvNTS (top row) and KcvS (bottom row) averaged over all F78 pairs in opposite (left column) and neighboring (right column) monomers. The geometric criteria are shown Figure S5. Numbers in the left panel denote different monomers. (b) Analogous data for KcvS S77G and KcvNTS G77S mutants, demonstrating the transfer of π-stack characteristics upon mutating the key residues. (c) Time series of the distances between the carbonyl oxygen of I73 and the hydroxyl hydrogen of S77 for all four monomers. (d) Schematic representation of a hydrogen bond formed between the amide oxygen of I73 and the hydroxyl hydrogen of S77 within monomer 3 of KcvS supplemented by orientation of F78 (snapshot taken at 80 ns). (e) Snapshots of F78 (red) in tetramer of KcvNTS (top) and KcvS (bottom) representing a wider and a narrow translocation pathway corresponding to statistics shown in a.