Abstract
Conformational changes in RNA play vital roles in the regulation of many biological systems, yet these changes can be challenging to visualize. Previously, we demonstrated that Pattern Recognition of RNA by Small Molecules (PRRSM) can unbiasedly cluster defined RNA secondary structure motifs utilizing an aminoglycoside receptor library. In this work, we demonstrate the power of this method to visualize changes in folding at the secondary structure level within two distinct riboswitch structures. After labeling at three independent positions on each riboswitch, PRRSM accurately classified all apo and ligand-bound riboswitch structures, including changes in the size of a structural motif, and revealed modification sites that prevented folding and/or led to a mixture of states. These data underscore the utility and robustness of the PRRSM assay for rapid assessment of RNA structural changes and for gaining ready insight into nucleotide positions critical to RNA folding.
Graphical abstract
INTRODUCTION
Noncoding RNA has been shown to adopt a multitude of roles in viral, bacterial, and mammalian systems.1 RNA structures are often responsive to binding partners or environmental conditions, and RNA conformational changes can be important in the modulation of cellular activities, especially gene expression.2–5 A range of current techniques allows structural insight into these changes, including NMR,6–16 fluorescence resonance energy transfer (FRET),17,18 single molecule force microscopy,19–22 and selective 2’-hydroxyl acylation by primer extension (SHAPE).23–27 While powerful, these techniques also suffer from limitations, including the time and/or quantity of sample needed for initial experiments. We proposed that our previously developed Pattern Recognition of RNA by Small Molecules (PRRSM) method could be used to provide rapid initial assessment of secondary-structure conformational changes with a single fluorophore label, as this technique requires minimal sample, no a priori structural insights, and simple instrumentation.
PRRSM has been shown to successfully classify RNA secondary structures utilizing an array of small molecule receptors.28 We have so far used a selection of 11 aminoglycoside receptors to distinguish the five canonical RNA motifs: bulges, hairpin loops, stems, symmetrical internal loops, and asymmetrical internal loops. We employ a fluorescence assay with the solvatochromic chemosensor, benzofuranyl uridine (BFU), which was initially incorporated into 16 RNA structures with variable sequences.28–34 The BFU modification is less bulky than other fluorophores, making it possible to modify more sites within compact RNA structures. Using the raw fluorescence data in principal component analysis (PCA), an unbiased statistical method, we were able to classify the five motifs with 100% predictive power and gained insight into the important topological differences that allowed structural differentiation. Recently, we extended the method to classify 13 individual sequences and differing secondary structure motif sizes, increasing the utility of PRRSM to determine the identity and nucleotide count of individual RNA secondary structures and sequences.35 This foundational work has shown PRRSM is an inexpensive, rapid throughput method for classifying simple RNA structure and for gaining insight into fundamental RNA: small molecule recognition principles, including the contributions of RNA topology and dynamics. Building upon this work, we herein demonstrate the utility of PRRSM to classify complex RNA structures and global conformation changes as well as to reveal critical tertiary interactions through perturbation to the native structure. These insights are achieved through the strategic placement of individual BFU modifications within a given RNA. As model systems, we employed multiple RNA constructs of BFU-modified pre-queuosine 1 riboswitch (PreQ1-RS) and fluoride riboswitch (F-RS) and analyzed the secondary structures in both the free and ligand-bound states.
RESULTS/DISCUSSION
Pre-Queuosine 1 Riboswitch.
To begin, we examined the Bacillus subtilis PreQ1-RS, which is an important regulator in the queuosine biosynthesis pathway through control of downstream queuosine biosynthesis genes.8,3–39 PreQ1-RS is a small 36 nucleotide (nt) riboswitch with an ~20 nM binding affinity for the prequeuosine 1 (PreQ1) ligand. The bound structure of the riboswitch is highly compact,10,38 and it was expected to test the limits of BFU chemosensor in the PRRSM assay.
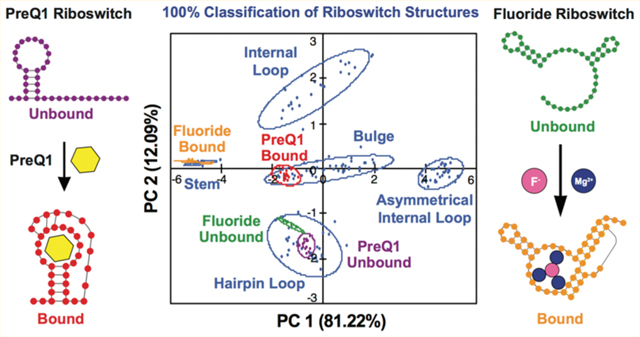
The PreQ1 unbound and bound states show several structural changes within the secondary structure motifs. In the unbound state, PreQ1-RS consists of a hairpin loop followed by a 12 nt single-stranded section; upon addition of PreQ1 ligand, the single-stranded region folds and forms base pairs with the hairpin loop, creating a smaller hairpin loop pocket for the PreQ1 ligand (Figure 1).8,36 Three uridines were chosen (U9, U11, and U14) to be replaced with BFU based on their structural transitions in the RNA upon binding of the PreQ1 ligand. In the unbound state, all modified uridines were within a 12 nt hairpin loop structure. Once in the bound state, U9 (PreQ1-BFU-9) is located in a 3 nt bulge motif, U11 (PreQ1-BFU-11) in a stem motif, and U14 (PreQ1-BFU-14) in a 4 nt hairpin loop as revealed by the solution structure (PDB 2L1V).8 As a control, we also labeled U22 (PreQ1-BFU-22), which is expected to remain in a stem structure in both the unbound and bound structures. After determining the uridine modification sites, we tested the four RNA constructs in the PRRSM assay.
Figure 1.
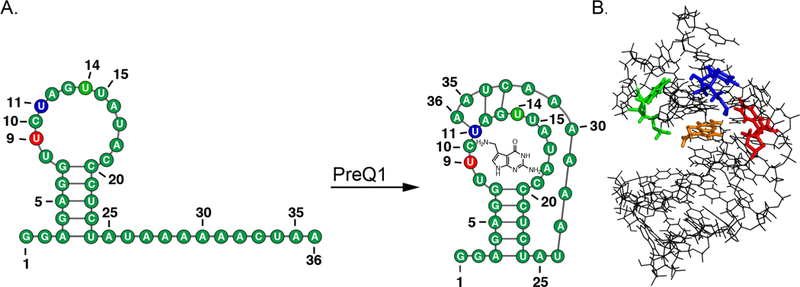
(A) PreQ1-RS riboswitch secondary structure in the unbound and bound state. (B) The bound tertiary structure of PreQ1-RS (PDB 2L1 V) with sites of BFU fluorophore insertion (U9-red, U11-blue, U14-green) and PreQ1 ligand (orange).8
We utilized both a standard ligand:RNA binding buffer ideal for separating motif classes (Condition (A), 10 mM NaH2PO4, 25 mM NaCl, 4 mM MgCl2, 0.5 mM ethylenediaminetetra-acetic acid (EDTA), pH 7.3 at 25 °C)28 and the same buffer with increased temperature and polyethylene glycol (PEG) added (Condition B, 10 mM NaH2PO4, 25 mM NaCl, 4 mM MgCl2, 0.5 mM EDTA, 8 mM PEG 12 000 pH 7.3 at 37 °C),35 which is known to denature secondary structures and stabilize tertiary structures, respectively.40 Condition (B) has previously been shown to allow separation of a range of individual nonstem secondary structure sequences and similar secondary structures with differing motif size.35
Under Condition (A), we found that the labeled nucleotides in all three experimental RNA constructs were correctly predicted by the PRRSM assay to be in hairpin loops in the absence of PreQ1 ligand (Figure 2, data shown in Figure S5–1). The control RNA construct (PreQ1-BFU-22) was also accurately predicted to be in a stem (Figure S1–1A). In this analysis, we start with the PCA plots previously determined for the RNA training set in which the axes represent a combination of variables best explaining the variance and covariance within the training set data. After defining the motif clusters of the training set, we input the data from the PreQ1-RS constructs and determined into which secondary structure motif class these constructs are most predicted to fall.
Figure 2.
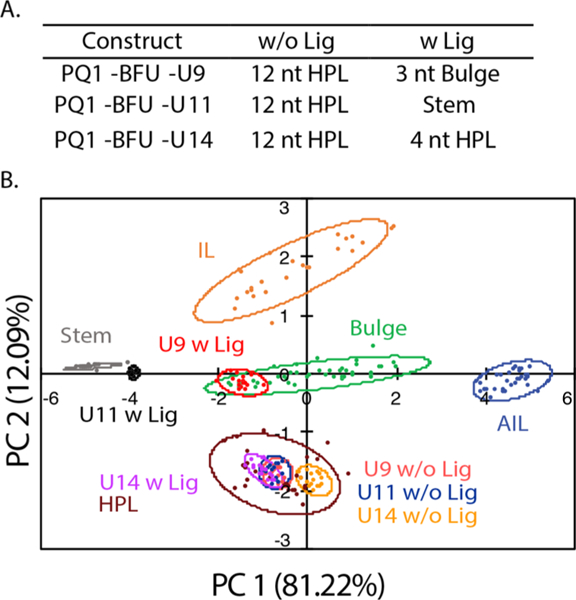
(A) The expected secondary structures of the PreQ1-RS constructs in the unbound (w/o Lig) and bound (w Lig) states. (B) PCA plot of the U9, U11, and U14 modified RNA in the absence (w/o Lig) and presence (w Lig) of PreQ1 ligand. All constructs were predicted to be the correct structure in both the unbound and bound states. Condition (A): 10 mM NaH2PO4, 25 mM NaCl, 4 mM MgCl2, 0.5 mM EDTA, pH 7.3 at 25 °C.
PreQ1 ligand was then titrated into 200 nM RNA for all three PreQ1-RS constructs. The BFU fluorophore caused a shift in the equilibrium of the ligand:riboswitch binding toward the apo state; therefore, a higher concentration of ligand was necessary to induce maximal binding compared to literature (Figure S1–2).36 For all four constructs, addition of 1.2 μM ligand (6 times the concentration of RNA) was utilized for the binding experiments. Upon addition of PreQ1 ligand, PreQ1-BFU-9, PreQ1-BFU-14, and PreQ1-BFU-U22 RNA constructs were predicted to adopt the expected secondary structure motifs (Figure 2 and Figure S5–1). While the PreQ1-BFU-U11 RNA construct did not fall within the 95% confidence interval predicted for stem structures by PRRSM, it was closely positioned to the stem cluster and was thus indeed predicted to be a stem structure through our standard of leave-one-out cross-validation (LOOCV). This difference could have been caused by the modified nucleotide being near the end of the stem, compared to the training set RNA stems, where the BFU is in the center of a stem structure. The central position is expected to increase the likelihood of a BFU to be base paired relative to a more flexible end position. Even with this difference, these results demonstrate that the PRRSM assay is able to correctly determine global conformational changes within complex RNA structures.
We then tested whether the size of the respective motifs, particularly the change in the hairpin loop size of the U14 label, could be detected by PRRSM using Condition B. It is important to note that, in previous work using Condition (B), individual stem sequences were not well-differentiated, and their inclusion negatively impacted the prediction of other secondary structures by PCA.35 As a result, they are not included in the PRSSM analysis with Condition (B), and therefore we did not include PreQ1-BFU-11 in this second analysis.
The PreQ1-BFU-9 and PreQ1-BFU-14 constructs were first tested based on the previous differentiation of individual sequences with Condition (B). In the unbound state, the PreQ1-RS hairpin loop is larger than the motifs in the training set, and thus both constructs were predicted to be the same size as the largest hairpin loop from the training set (six nucleotides). Future work will thus be focused on expanding the size of the motifs within the training set.
Upon addition of ligand, PreQ1-BFU-9 is correctly determined to be in a three-nucleotide bulge. Even more notably, the shift of PreQ1-BFU-14 from a 12 nt hairpin loop to a 4 nt hairpin loop in the bound state could be accurately predicted via PRRSM (Figure S1–3, data shown in Figure S5–2). These data underscore the utility and robustness of the PRRSM assay by demonstrating its ability to distinguish not only conformational changes but also motif size in unbound and bound states. Additionally, these data are consistent with minimal perturbation of riboswitch conformations by the BFU label, which was further confirmed by NMR (Figure S2–1,2,3). PRRSM was able to classify all preQ1-RS RNA constructs with 100% accuracy, showing the effectiveness of this technique in analyzing riboswitch structural conformational changes.
Fluoride Riboswitch.
To further evaluate the generality of the PRRSM assay, we analyzed conformational changes in the Bacillus cereus fluoride riboswitch (F-RS; Figure 3).16,41 F-RS regulates gene expression for fluoride toxicity response genes and is widespread in bacteria and archaea.5 Specifically, F-RS upregulates the transcription of fluoride transporters that facilitate expulsion of fluoride from the cell.5 In the presence of fluoride and magnesium, the riboswitch forms a tight binding site for the fluoride anion encapsulated by three magnesium cations, which was revealed by the crystal structure of the fluoride-bound Thermotoga petrophila fluoride riboswitch.41 Recently, it has been shown that the B. cereus F-RS folds into essentially identical structures in the presence of magnesium regardless of the presence of fluoride, wherein the magnesium-bound state undergoes a distinct fleeting dynamic process and has a reduced stability compared to the fluoride-bound state.16 In the absence of magnesium, the B. cereus F-RS is largely unfolded, only forming one 4 nt loop hairpin and one 6 nt loop hairpin structures, and undergoes a large conformational change upon ligand binding (Figure 3). This complex ligand-dependent conformational transition,16 together with a highly compact ligand binding pocket,41 makes the F-RS an interesting model for characterizing RNA conformational change using PRRSM. Thus, we studied conformational changes of the B. cereus F-RS.12
Figure 3.
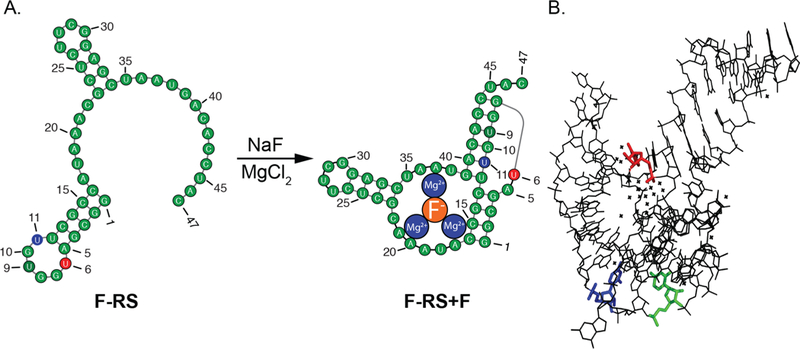
(A) B. cereus fluoride riboswitch in the unbound (F-RS) and bound (F-RS + F) states. (B) Tertiary structure of T. petrophila Fluoride riboswitch (PDB 4ENC), which folds into a nearly identical structure of B. cereus F-RS, with the three BFU modification sites, F-RS-BFU-6 (red), F-RS-BFU-9 (green), and F-RS-BFU-11 (blue).
Within the F-RS structure, we decided to modify three uridine sites (F-RS-BFU-6, −9, and −11) located in the 6 nt hairpin loop of the unfolded riboswitch. U9 and U11 sites transition from the loop structure into a stem structure in the bound state, while U6 nucleotide transitions to a 2 nt bulge motif. As a U6-to-C6 mutation was previously found to be deleterious to folding, the BFU-6 modification was designed to serve as an additional control on the ability of BFU-labeled structures to recapitulate established conformations.16 In this case, the BFU-6 label was expected to similarly disrupt folding of the F-RS, and thus the FR-BFU-U6 construct would demonstrate no change in structure in the presence of fluoride. We again selected a control RNA construct labeled at a position not expected to change upon riboswitch folding, in this case U25 (F-RS-BFU-25). The fluoride riboswitch constructs were tested in the PRRSM assay in the absence and presence of fluoride utilizing a standard ligand:RNA binding buffer ideal for separating motif classes, Condition (A) (10 mM NaH2PO4, 25 mM NaCl, 4 mM MgCl2, 0.5 mM EDTA, pH 7.3 at 25 °C).28,35 Interestingly, F-RS-BFU-6, −9, and −11 in the unbound states were determined to be in a hairpin loop even in the presence of magnesium (Figure 4, data shown in Figure S5–3). Since the magnesium-bound pseudoknot structure of F-RS is metastable,14 these results suggest that BFU modification may introduce subtle steric effects on the compact structure, shifting the conformational equilibrium toward the extended magnesium-free hairpin loop structure12 in the absence of fluoride. We then used saturating conditions of fluoride (10 mM) to determine the classification of the F-RS bound structures.14 Upon the addition of fluoride, the F-RS-BFU-6 showed little change in clustering between the unbound and bound states, consistent with the published mutation studies.16 On the other hand, F-RS-BFU-9 was successfully classified into the correct folded conformation, and F-RS-BFU-11 was determined to be folding to a stem structure only ~50% of the time. The F-RS-BFU-11 modification is positioned near the end of a stem motif in the pseudoknot, suggesting that either the receptors interacted differently with this region as compared to the more rigid central stem position or the modification impacts the folding of the riboswitch. F-RS-BFU-25 was accurately predicted to be in a stem in both the unbound and bound forms. (Figure S1–1B).
Figure 4.
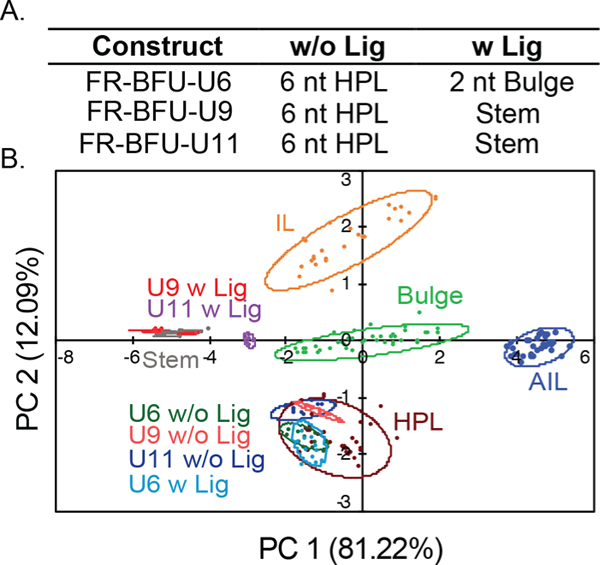
(A) The expected secondary structures of the F-RS constructs in the unbound (w/o Lig) and bound (w Lig) states. (B) PCA plot of the U9 and U11 modified RNA in the absence (w/o Lig) and presence (w Lig) of fluoride. U9 was correctly determined, while U11 was not predicted to be a stem structure in the bound state. F-RS-BFU-11 was positioned near the end of a stem and thus may interact differently with the aminoglycoside receptors compared to central stem modification. Condition (A): 10 mM NaH2PO4, 25 mM NaCl, 4 mM MgCl2, 0.5 mM EDTA, pH 7.3 at 25 °C.
Given that clustering of F-RS-BFU-11 “bound” state was found to be between the stem and hairpin loop training set clusters, we first tested the hypothesis that both the bound and unbound states are present and represented. Accordingly, we subjected differing ratios of representative stem and hairpin loop structures to the PRRSM assay. While keeping the total RNA concentration at 200 nM, the relative percentage of two RNA constructs was shifted from 100% stem to 100% hairpin loop in 25% increments. At either 75% stem or hairpin loop, there is a minimal shift from the 95% confidence intervals of the corresponding constructs (Figure S1–4). When the two constructs are at equal concentrations, the RNA clusters shift near stem, hairpin loop, and bulge clusters. Similar trends were seen with the F-RS-BFU-11 construct in the presence of fluoride. These results are consistent with the hypothesis that both the unbound and bound states are present and suggest a change in the conformational equilibrium, though a uniform misfolded structure cannot be ruled out.
To examine the potential effect of BFU modification on conformational equilibrium of the F-RS, we further performed 1H imino NMR titration studies of all three modified F-RS constructs. Each RNA construct was initially titrated with magnesium from 0 to 5 mM, followed by titration of fluoride from 0 to 5 mM at both 10 and 30 °C. The folding and binding ability of the BFU-modified constructs were then compared to the unmodified riboswitch. The F-RS-BFU-6 construct displays no binding to either magnesium or fluoride under the measured conditions, consistent with the PRRSM assay results and previous mutation studies (Figure S2–4). This result further suggests PRRSM may be a tool to visualize modifications that perturb folding interactions in unknown RNA structures.
The NMR spectra of F-RS-BFU-9 showed the expected conformational changes upon the addition of magnesium and fluoride, though higher concentrations of ligand were required (Figure 5). In addition, the broadened NMR spectrum of F-RS-BFU-9 in the presence of 5 mM magnesium indicates that the unbound F-RS-BFU-9 does not adopt the well-folded pseudoknot of the unmodified riboswitch, which is also consistent with the PRRSM results. The final RNA construct, F-RS-BFU-11, had reduced magnesium and fluoride binding compared to the unmodified construct. The BFU modification at the U11 site was likely positioned within the binding pocket, impacting the ability of magnesium to bind in the less stable prefolded structure. With addition of the fluoride anion, the equilibrium was shifted significantly to the bound state at saturating conditions. PRRSM was thus able to accurately classify 80% of the F-RS constructs, and reduced classification was confirmed to be the result of perturbation by BFU of native contacts. It is important to note that the PRRSM assay is an ensemble measurement, and, while it is not able to distinguish two coexisting structures, our results demonstrate that it does reveal the occurrence of multiple structures. PRRSM thus can serve as a simple and rapid technique to determine if RNA sequences, including modified RNA sequences, are able to switch conformations.
Figure 5.
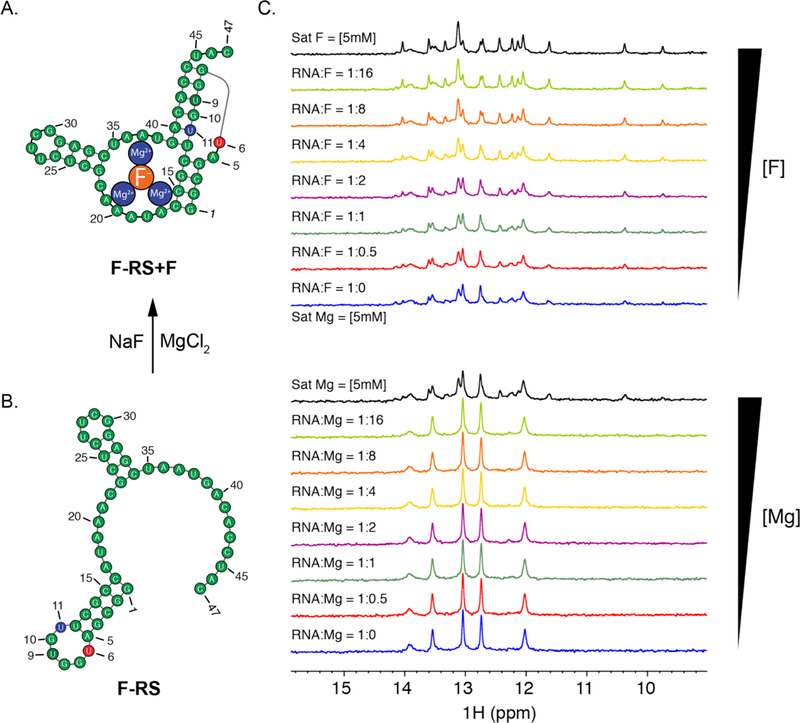
B. cereus fluoride riboswitch in the bound (A) and unbound (B) states. (C) 1D 1H imino NMR of F-RS-BFU-U9 from 0 to 5 Mm magnesium followed by 0−5 mMfluoride at 30 °C. Titration of magnesium shifted the peaks, suggesting a change of the equilibrium between the unbound and bound states. Upon addition of fluoride, there was a complete shift to the bound state. Similar results were seen at 10 °C. The F-RS-BFU-U6 and U11 1D 1H imino NMR are shown in the Supporting Information (Figure S2–S4,5,6). Buffer conditions: 10 mM NaH2PO4, 50 mM KCl, and 50 μM EDTA, pH 6.4.
Given literature evidence that some small molecules can shift the conformational equilibrium of RNA,42–44 we took advantage of the fluoride riboswitch system to examine the potential effect of our aminoglycoside receptors on RNA conformational equilibrium and therefore the PRRSM clustering. Utilizing the one-dimensional (1D) 1H imino proton NMR measurements, we analyzed the conformations of F-RS-BFU-9 in the presence of two aminoglycosides, namely, apramycin and kanamycin (Figure 6). Based on fluorescence changes with these aminoglycosides, we expected apramycin, which demonstrates significant differences in fluorescence between the unbound and bound states of the F-RS-BFU-9 construct, to serve as a positive binding control and kanamycin, which showed little fluorescence change between the two states, to serve as a negative binding control. The equilibrium of the RNA construct characterized by NMR was unaffected by either aminoglycoside receptor at both 10 and 30 °C. Importantly, these data further support the versatility and robustness of the PRRSM assay toward more complex RNA by demonstrating that any changes in RNA secondary structure clustering are the direct result of RNA folding and not interference by the receptors.
Figure 6.
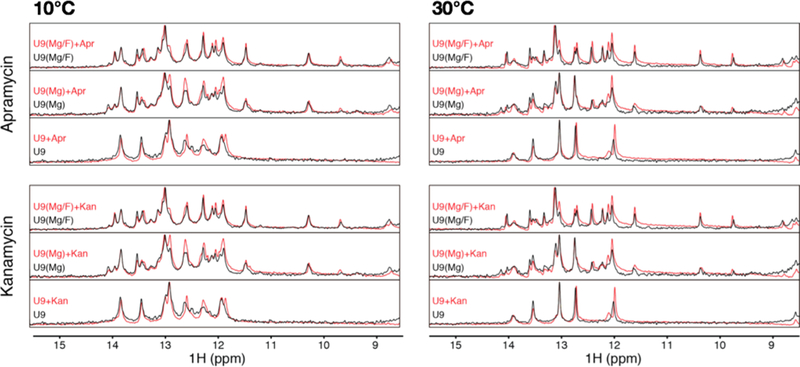
1D 1H imino proton NMR of F-RS-BFU-U9 (0.1 mM) in the presence of 0.5 mM apramycin or kanamycin. Addition of either receptor showed no change at both 10 and 30 °C. Buffer conditions: 10 mM NaH2PO4, 50 mM KCl, and 50 μM EDTA, pH 6.4.
CONCLUSIONS
In conclusion, we have demonstrated the ability of the PRRSM assay to distinguish multiple complex RNA structures across ligand-induced conformational changes. The BFU fluorophore, when positioned away from the ligand binding pocket, minimally affects RNA structures with conformational changes. Importantly, the PRRSM assay accurately reported any impact of these modifications, which were confirmed using 1D NMR. PRRSM is thus an orthogonal technique that adds to the existing repertoire of RNA structural and conformational interrogation methods by enabling the rapid classification of highly flexible RNA secondary structures and potentially revealing critical interaction sites in the folded structure. We note that labeling an RNA construct at multiple positions, as performed here, will be important for the determination of unknown RNA structure changes. Subsequent experiments will focus on expanding the RNA training set, which will both enhance the current classification ability of PRRSM and provide additional insight into RNA:small molecule interactions, and the screening of alternative or inhibitory riboswitch ligands that introduce structural changes. Future work will focus on transitioning PRRSM to more sophisticated machine-based learning techniques, investigating conformational switching in other disease-relevant RNAs such as the regulatory RNA elements in HIV,45 and utilizing PRRSM to ascertain classification of RNA tertiary structures.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge J. Forte for synthesis of the BFU chemosensor, and the laboratory of H. Al-Hashimi for access to the MerMade oligonucleotide synthesizer. We thank the members of the Hargrove lab for stimulating discussion and input as well as G. Young for maintenance of NMR instruments at the Univ. of North Carolina at Chapel Hill. A.E.H. wishes to acknowledge financial support for this work from Duke Univ., the National Institute of Health (U54GM103297), and the Research Corporation for Science Advancement Cottrell Scholar Award. C.S.E. was supported in part through U.S. Department of Education GAANN Fellowship (P200A150114). Q.Z. wishes to acknowledge start-up funding from the Univ. of North Carolina at Chapel Hill and the National Institute of Health (R01 GM114432).
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.8b09665.
Riboswitch principal component analysis, NMR titration of fluoride and PreQ1 riboswitches, RNA solid-phase synthesis and purification, materials and synthetic methods, aminoglycoside receptor: riboswitch titration graphs, additional references (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Cech TR; Steitz JA The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014, 157 (1), 77–94. [DOI] [PubMed] [Google Scholar]
- (2).Soukup JK; Soukup GA Riboswitches exert genetic control through metabolite-induced conformational change. Curr. Opin. Struct. Biol. 2004, 14 (3), 344–349. [DOI] [PubMed] [Google Scholar]
- (3).Breaker RR Prospects for riboswitch discovery and analysis. Mol. Cell 2011, 43 (6), 867–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Breaker RR Riboswitches and the RNA world. Cold Spring Harbor Perspect. Biol. 2012, 4 (2), No. a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Baker JL; Sudarsan N; Weinberg Z; Roth A; Stockbridge RB; Breaker RR Widespread Genetic Switches and Toxicity Resistance Proteins for Fluoride. Science 2012, 335 (6065), 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Edwards TE; Klein DJ; Ferre-D’Amare AR Riboswitches: small-molecule recognition by gene regulatory RNAs. Curr. Opin. Struct. Biol. 2007, 17 (3), 273–9. [DOI] [PubMed] [Google Scholar]
- (7).Buck J; Furtig B; Noeske J; Wohnert J; Schwalbe H Time-resolved NMR spectroscopy: ligand-induced refolding of ribos-witches. Methods Mol. Biol. (N. Y, NY, U. S.) 2009, 540, 161–71. [DOI] [PubMed] [Google Scholar]
- (8).Kang M; Peterson R; Feigon J Structural Insights into riboswitch control of the biosynthesis of queuosine, a modified nucleotide found in the anticodon of tRNA. Mol. Cell 2009, 33 (6), 784–90. [DOI] [PubMed] [Google Scholar]
- (9).Duchardt-Ferner E; Weigand JE; Ohlenschlager O; Schmidtke SR; Suess B; Wohnert J Highly modular structure and ligand binding by conformational capture in a minimalistic riboswitch. Angew. Chem., Int. Ed. 2010, 49 (35), 6216–9. [DOI] [PubMed] [Google Scholar]
- (10).Serganov A; Patel DJ Metabolite Recognition Principles and Molecular Mechanisms Underlying Riboswitch Function. Annu. Rev. Biophys. 2012, 41 (1), 343–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Reining A; Nozinovic S; Schlepckow K; Buhr F; Furtig B; Schwalbe H Three-state mechanism couples ligand and temperature sensing in riboswitches. Nature 2013, 499 (7458), 355–9. [DOI] [PubMed] [Google Scholar]
- (12).Zhao B; Hansen AL; Zhang Q Characterizing Slow Chemical Exchange in Nucleic Acids by Carbon CEST and Low Spin-Lock Field R1p NMR Spectroscopy. J. Am. Chem. Soc. 2014, 136 (1), 20–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Kang M; Eichhorn CD; Feigon J Structural determinants for ligand capture by a class II preQ1 riboswitch. Proc. Natl. Acad. Sci. U. S. A. 2014, 111 (6), E663–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Zhao B; Zhang Q Measuring Residual Dipolar Couplings in Excited Conformational States of Nucleic Acids by CEST NMR Spectroscopy. J. Am. Chem. Soc. 2015, 137 (42), 13480–13483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Ren A; Xue Y; Peselis A; Serganov A; Al-Hashimi HM; Patel DJ Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch. Cell Rep. 2015, 13 (9), 1800–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Zhao B; Guffy SL; Williams B; Zhang Q An excited state underlies gene regulation of a transcriptional riboswitch. Nat. Chem. Biol. 2017, 13, 968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Klostermeier D; Millar DP RNA Conformation and Folding Studied with Fluorescence Resonance Energy Transfer. Methods 2001, 23 (3), 240–254. [DOI] [PubMed] [Google Scholar]
- (18).Harbaugh S; Kelley-Loughnane N; Davidson M; Narayanan L; Trott S; Chushak YG; Stone MO FRET-based optical assay for monitoring riboswitch activation. Biomacromolecules 2009, 10 (5), 1055–60. [DOI] [PubMed] [Google Scholar]
- (19).Greenleaf WJ; Frieda KL; Foster DAN; Woodside MT; Block SM Direct Observation of Hierarchical Folding in Single Riboswitch Aptamers. Science 2008, 319 (5863), 630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Neupane K; Yu H; Foster DAN; Wang F; Woodside MT Single-molecule force spectroscopy of the add adenine riboswitch relates folding to regulatory mechanism. Nucleic Acids Res. 2011, 39 (17), 7677–7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Heus HA; Puchner EM; van Vugt-Jonker AJ; Zimmermann JL; Gaub HE Atomic force microscope-based single-molecule force spectroscopy of RNA unfolding. Anal. Biochem. 2011, 414 (1), 1–6. [DOI] [PubMed] [Google Scholar]
- (22).Frieda KL; Block SM Direct observation of cotranscriptional folding in an adenine riboswitch. Science 2012, 338 (6105), 397–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Lu C; Ding F; Chowdhury A; Pradhan V; Tomsic J; Holmes WM; Henkin TM; Ke A SAM Recognition and Conformational Switching Mechanism in the Bacillus subtilis yitJ S Box/SAM-I Riboswitch. J. Mol Biol. 2010, 404 (5), 803–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Hajdin CE; Bellaousov S; Huggins W; Leonard CW; Mathews DH; Weeks KM Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (14), 5498–5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Tyrrell J; McGinnis JL; Weeks KM; Pielak GJ The Cellular Environment Stabilizes Adenine Riboswitch RNA Structure. Biochemistry 2013, 52 (48), 8777–8785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Rice GM; Leonard CW; Weeks KM RNA secondary structure modeling at consistent high accuracy using differential SHAPE. RNA 2014, 20 (6), 846–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Watters KE; Yu AM; Strobel EJ; Settle AH; Lucks JB Characterizing RNA structures in vitro and in vivo with selective 2’-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq). Methods 2016, 103, 34–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Eubanks CS; Forte JE; Kapral GJ; Hargrove AE Small Molecule-Based Pattern Recognition To Classify RNA Structure. J. Am. Chem. Soc. 2017, 139 (1), 409–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Efthymiou T; Krishnamurthy R Microwave-Assisted Phosphitylation of DNA and RNA Nucleosides and Their Analogs In Current Protocols in Nucleic Acid Chemistry; John Wiley & Sons, Inc., 2001. [DOI] [PubMed] [Google Scholar]
- (30).Tanpure AA; Srivatsan SG Synthesis and Photophysical Characterisation of a Fluorescent Nucleoside Analogue that Signals the Presence of an Abasic Site in RNA. ChemBioChem 2012, 13 (16), 2392–2399. [DOI] [PubMed] [Google Scholar]
- (31).Ghanty U; Fostvedt E; Valenzuela R; Beal PA; Burrows CJ Promiscuous 8-Alkoxyadenosines in the Guide Strand of an SiRNA: Modulation of Silencing Efficacy and Off-Pathway Protein Binding. J. Am. Chem. Soc. 2012, 134 (42), 17643–17652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Gallagher-Duval S; Herve G; Sartori G; Enderlin G; Len C Improved microwave-assisted ligand-free Suzuki-Miyaura cross-coupling of 5-iodo-2[prime or minute]-deoxyuridine in pure water. NewJ. Chem. 2013, 37 (7), 1989–1995. [Google Scholar]
- (33).Meher G; Efthymiou T; Stoop M; Krishnamurthy R Microwave-assisted preparation of nucleoside-phosphoramidites. Chem. Commun. 2014, 50 (56), 7463–7465. [DOI] [PubMed] [Google Scholar]
- (34).Tanpure AA; Srivatsan SG Conformation-sensitive nucleoside analogues as topology-specific fluorescence turn-on probes for DNA and RNA G-quadruplexes. Nucleic Acids Res. 2015, 43 (22), No. e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Eubanks CS; Hargrove AE Sensing the impact of environment on small molecule differentiation of RNA sequences. Chem. Commun. 2017, 53 (100), 13363–13366. [DOI] [PubMed] [Google Scholar]
- (36).Roth A; Winkler WC; Regulski EE; Lee BW; Lim J; Jona I; Barrick JE; Ritwik A; Kim JN; Welz R; Iwata-Reuyl D; Breaker RR A riboswitch selective for the queuosine precursor preQ1 contains an unusually small aptamer domain. Nat. Struct. Mol. Biol. 2007, 14 (4), 308–17. [DOI] [PubMed] [Google Scholar]
- (37).Klein DJ; Edwards TE; Ferré-D’Amaré AR Cocrystal structure of a class I preQ1 riboswitch reveals a pseudoknot recognizing an essential hypermodified nucleobase. Nat. Struct. Mol. Biol. 2009, 16 (3), 343–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Jenkins JL; Krucinska J; McCarty RM; Bandarian V; Wedekind JE Comparison of a preQ1 riboswitch aptamer in metabolite-bound and free states with implications for gene regulation. J. Biol. Chem. 2011, 286 (28), 24626–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Zhang Q; Kang M; Peterson RD; Feigon J Comparison of Solution and Crystal Structures of PreQ1 Riboswitch Reveals Calcium-Induced Changes in Conformation and Dynamics. J. Am. Chem. Soc. 2011, 133 (14), 5190–5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Tyrrell J; Weeks KM; Pielak GJ Challenge of mimicking the influences of the cellular environment on RNA structure by PEG-induced macromolecular crowding. Biochemistry 2015, 54 (42), 6447–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Ren A; Rajashankar KR; Patel DJ Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch. Nature 2012, 486 (7401), 85–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Bardaro JMF; Shajani Z; Patora-Komisarska K; Robinson JA; Varani G How binding of small molecule and peptide ligands to HIV-1 TAR alters the RNA motional landscape. Nucleic Acids Res. 2009, 37 (5), 1529–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Stelzer A; Frank A; Kratz J; Swanson M; Gonzalez-Hernandez M; Lee J; Andricioaei I; Markovitz D; Al-Hashimi H Discovery of selective bioactive small molecules by targeting an RNA dynamic ensemble. Nat. Chem. Biol. 2011, 7 (8), 553–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Rizvi NF; Howe JA; Nahvi A; Klein DJ; Fischmann TO; Kim H-Y; McCoy MA; Walker SS; Hruza A; Richards MP; Chamberlin C; Saradjian P; Butko MT; Mercado G; Burchard J; Strickland C; Dandliker PJ; Smith GF; Nickbarg EB Discovery of Selective RNA-Binding Small Molecules by Affinity- Selection Mass Spectrometry. ACS Chem. Biol. 2018, 13 (3), 820–831. [DOI] [PubMed] [Google Scholar]
- (45).Keane SC; Summers MF NMR Studies of the Structure and Function of the HIV-1 5 ‘-Leader. Viruses 2016, 8 (12), 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


