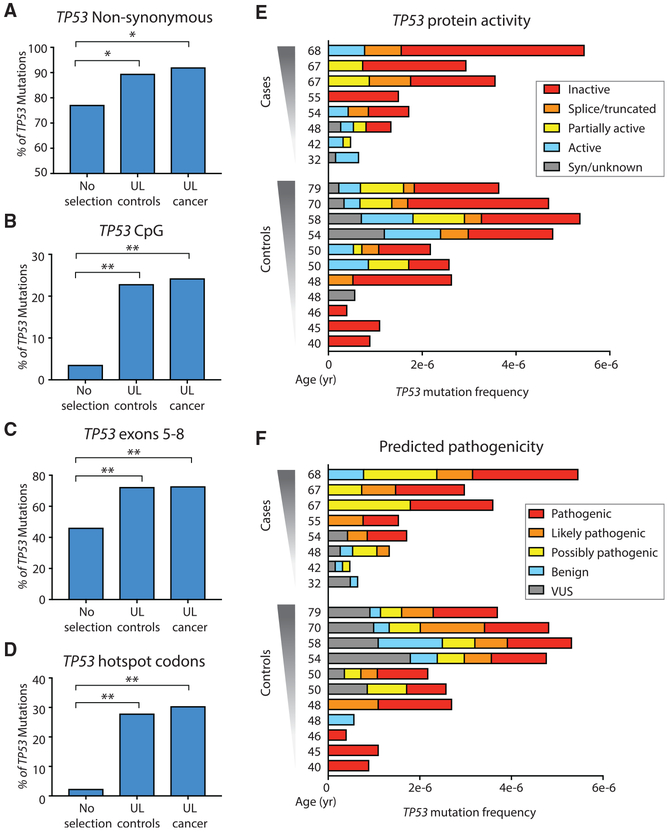
Figure 3. Evidence of Positive Selection in TP53 Background Mutations from ULs.
(A–D) Percentage of non-synonymous TP53 mutations (A), and percentage of TP53 mutations localized in CpG dinucleotides (B), in exons 5–8 (C), and in hotspot codons (D). For (A–D), TP53 mutations identified in UL from controls and cancer significantly exceed expected values without selection. *p < 0.05, **p < 0.0001 by binomial exact test, n = 79 for UL controls, and n = 33 for UL cancer. (E and F) Protein activity (E) and predicted pathogenicity (F) color-coded as five groups from Seshat data. Patients are sorted by ascending age. For each patient, TP53 mutation frequency is calculated as the number of mutations in the coding region divided by the total number of DS nucleotides sequenced in that region. Two cancer patients with unusually low sequencing depth and no identified TP53 mutations are not shown. Nearly all cases and controls carry mutations that have an impact on protein activity and predicted pathogenicity.