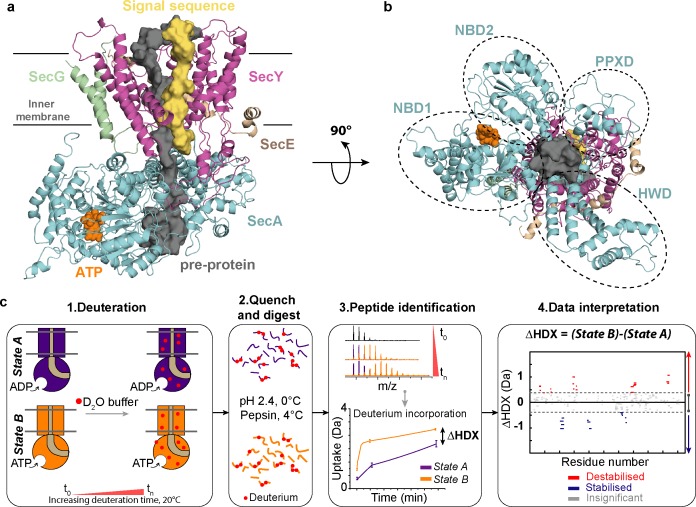
Figure 1. Structure of SecA-SecYEG complex and HDX-MS workflow.
(a) Structure and sub-domains of SecA-SecYEG (based in 3DIN as in Zimmer et al., 2008, modelled with a pre-protein; Corey et al., 2016a). The pre-protein, ATP and signal sequence are highlighted in grey, orange and yellow, respectively. (b) Top view of the complex highlighting the nucleotide-binding-domains NBD1 and NBD2 as well as PPXD (Pre-Protein cross-linking Domain) and HWD (Helical Wing Domain) (c) Overview of the HDX-MS process. The sample is prepared in detergent micelles (DDM) and after addition of nucleotides is incubated in a deuterated solvent (1). Following deuteration of the mixture at different time-points, the HDX reaction is quenched and the protein is digested with pepsin (2). Peptides are separated by liquid chromatography and subsequently identified by mass spectrometry (MS). The mass uptake of the protein in different conditions (e.g. AMPPNP vs ADP) is then compared (3). Peptides with significant difference in deuterium uptake are mapped onto a Woods plot (4). The length of lines represent the peptide sizes. Blue and red regions indicate significant protection and deprotection, respectively. Insignificant differences, calculated by a 99% confidence interval, are shown in grey.