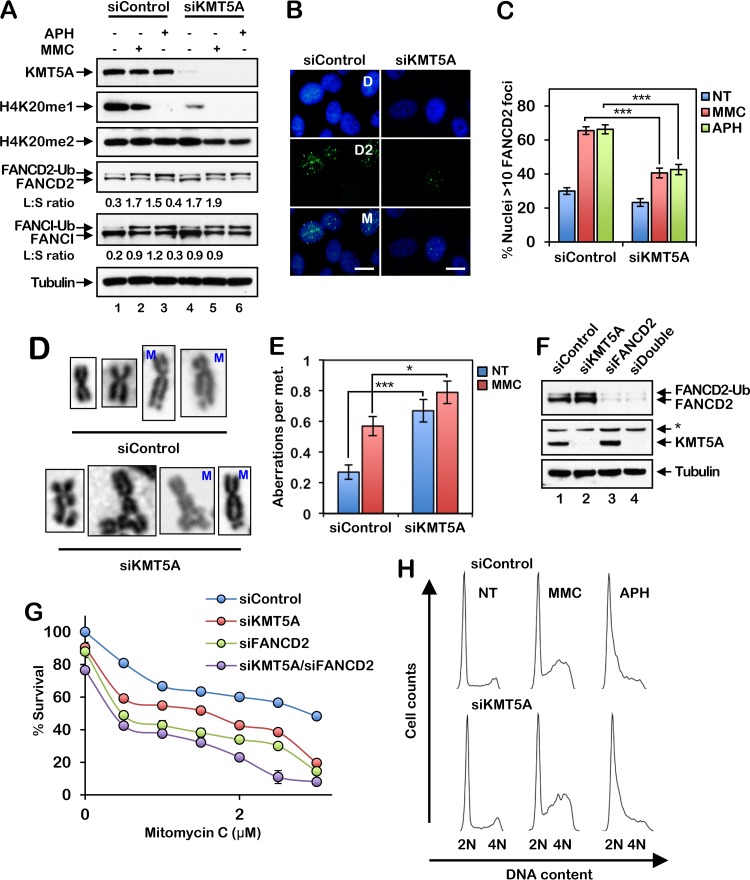
FIG 2.
The histone methyltransferase KMT5A is necessary for efficient activation of the FA pathway and ICL repair. (A) HeLa cells were incubated with control nontargeting siRNA (siControl) or siRNA targeting KMT5A (siKMT5A) for 64 h. The cells were then treated with 200 nM mitomycin C (MMC) or 1 μM aphidicolin (APH) for 16 h, and whole-cell lysates were analyzed by immunoblotting. Ub, ubiquitin; L:S ratio, ratio of monoubiquitinated to nonubiquitinated FANCD2 or FANCI. (B) Representative images of FANCD2 foci in MMC-treated HeLa cells. Cells were treated as described in the legend to panel A. D, DAPI; D2; FANCD2; M, merge. Bars, 10 μm. (C) Quantification of FANCD2 focus formation for the assay whose results are presented in panel B. Nuclei with >10 FANCD2 foci were considered positive. At least 300 nuclei were scored per biological replicate. (D and E) HeLa cells were incubated with siControl or siKMT5A for 64 h. Cells were then treated with 10 nM MMC for 16 h, and metaphase (met.) chromosomes were analyzed for the presence of structural aberrations. Representative chromosome images are shown in panel D. M, mitomycin C. Fifty metaphases were scored per treatment. These experiments were performed twice with similar results. Error bars represent the standard errors of the means. *, P < 0.05; ***, P < 0.001. (F) HeLa cells were incubated with siControl, siKMT5A, siFANCD2, or siKMT5A plus siFANCD2 (siDouble) for 64 h, and whole-cell lysates were analyzed by immunoblotting. *, nonspecific band. (G) Cells were treated with increasing concentrations of MMC for 48 h, and percent survival was determined using an 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium survival assay. Error bars represent the standard errors of the means from four replicates. Experiments were performed twice with similar results. (H) HeLa cells were incubated with siControl or siKMT5A for 64 h and then treated with 200 nM MMC or 1 μM APH for 16 h. Cells were fixed, stained with propidium iodide, and analyzed by flow cytometry. Cell cycle analysis was performed using FlowJo (v10.2) software.