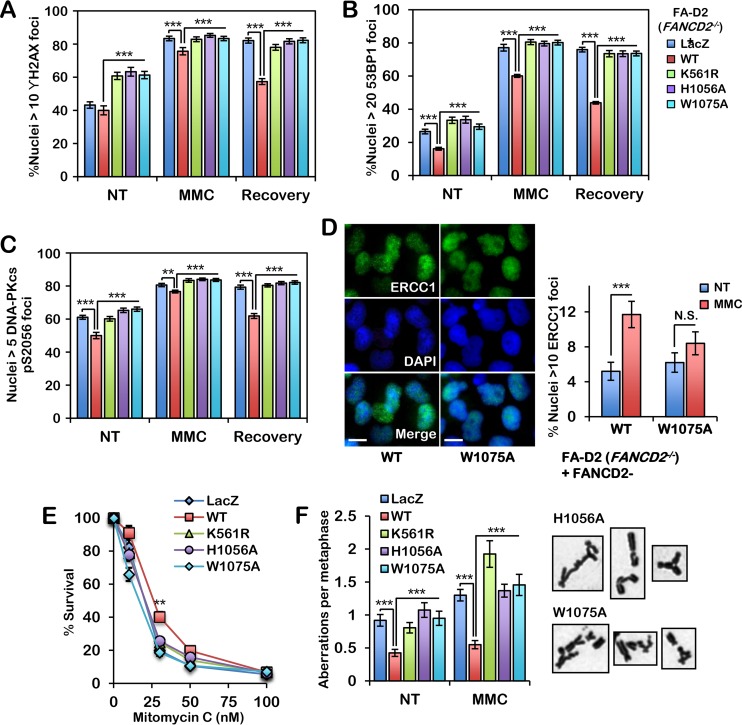
FIG 5.
The FANCD2 HBD/MBD is required for efficient conservative ICL repair. (A to C) FA-D2 cells stably expressing LacZ, wild-type FANCD2, FANCD2-K561R, FANCD2-H1056A, and FANCD2-W1075A were incubated in the absence or presence of 200 nM mitomycin C (MMC) for 24 h and allowed to recover for an additional 24 h. Cells were fixed and stained with anti-γH2AX (A), anti-53BP1 (B), or anti-DNA-PKcs pS2056 (C) antibodies, and the numbers of nuclei with >10 γH2AX (A), >20 53BP1 (B), or >5 DNA-PKcs pS2056 (C) foci were scored. At least 300 nuclei were scored per biological replicate. (D) FA-D2 cells stably expressing wild-type FANCD2 or FANCD2-W1075A were incubated in the absence (not treated [NT]) or presence of 200 nM MMC for 24 h. Cells were fixed and stained with anti-ERCC1, and the numbers of nuclei with >10 large foci were scored. At least 300 nuclei were scored per sample. Experiments were performed twice with similar results. Error bars represent the standard errors of the means from two independent experiments. ***, P < 0.001; N.S., not significant. (E) The same cells used in the assay whose results are presented in panels A to C were incubated in the presence of various concentrations of MMC for 7 to 10 days, the surviving cells were stained with crystal violet, and percent survival relative to the number of surviving untreated cells was scored. (F) The same cells were incubated in the absence (nontreated) or presence of 16 nM MMC for 24 h, and metaphase chromosomes were analyzed for structural aberrations, including gaps, breaks, and radial formations. Representative chromosome aberrations from MMC-treated FA-D2 cells expressing FANCD2-H1056A and FANCD2-W1075A are shown. All experiments were performed three times with similar results, except for those whose results are presented in panel F, which were performed twice. Error bars represent the standard errors of the means. For the assay whose results are presented in panel F, 80 metaphases were scored per treatment. **, P < 0.01; ***, P < 0.001.