Mx proteins are evolutionarily conserved in vertebrates and can restrict a wide range of viruses in a cell-autonomous way. The contribution to antiviral defense of Mx1 expression in hematopoietic cells remains largely unknown. We show that protection against influenza virus infection requires Mx1 expression in the nonhematopoietic cellular compartment. In contrast, Mx1 in bone marrow-derived cells is sufficient to control disease and virus replication following infection with a Thogoto virus. This indicates that, in addition to its well-established antiviral activity in nonhematopoietic cells, Mx1 in hematopoietic cells can also play an important antiviral function. In addition, cells of hematopoietic origin that lack a functional Mx1 gene contribute to Thogoto virus dissemination and associated disease.
KEYWORDS: bone marrow chimeras, Mx1, Thogoto virus, myeloid cells
ABSTRACT
Myxovirus resistance 1 (Mx1) is an interferon-induced gene that encodes a GTPase that plays an important role in the defense of mammalian cells against influenza A and other viruses. The Mx1 protein can restrict a number of viruses independently of the expression of other interferon-induced genes. Mx genes are therefore considered to be an important part of the innate antiviral immune response. However, the possible impact of Mx expression in the hematopoietic cellular compartment has not been investigated in detail in the course of a viral infection. To address this, we performed bone marrow chimera experiments using congenic B6.A2G Mx1+/+ and B6.A2G Mx1−/− mice to study the effect of Mx1 expression in cells of hematopoietic versus nonhematopoietic origin. Mx1+/+ mice were protected and Mx1−/− mice were susceptible to influenza A virus challenge infection, regardless of the type of bone marrow cells (Mx1+/+ or Mx1−/−) the animals had received. Infection with Thogoto virus, however, revealed that Mx1−/− mice with a functional Mx1 gene in the bone marrow compartment showed reduced liver pathology compared with Mx1−/− mice that had been grafted with Mx1−/− bone marrow. The reduced pathology in these mice was associated with a reduction in Thogoto virus titers in the spleen, lung, and serum. Moreover, Mx1+/+ mice with Mx1−/− bone marrow failed to control Thogoto virus replication in the spleen. Mx1 in the hematopoietic cellular compartment thus contributes to protection against Thogoto virus infection.
IMPORTANCE Mx proteins are evolutionarily conserved in vertebrates and can restrict a wide range of viruses in a cell-autonomous way. The contribution to antiviral defense of Mx1 expression in hematopoietic cells remains largely unknown. We show that protection against influenza virus infection requires Mx1 expression in the nonhematopoietic cellular compartment. In contrast, Mx1 in bone marrow-derived cells is sufficient to control disease and virus replication following infection with a Thogoto virus. This indicates that, in addition to its well-established antiviral activity in nonhematopoietic cells, Mx1 in hematopoietic cells can also play an important antiviral function. In addition, cells of hematopoietic origin that lack a functional Mx1 gene contribute to Thogoto virus dissemination and associated disease.
INTRODUCTION
Myxovirus resistance proteins are dynamin-like large GTPases that can inhibit a wide array of viruses, including members of the Orthomyxoviridae, Rhabdoviridae, and Bunyaviridae (1). Mx genes are evolutionarily conserved in vertebrates, and their expression is induced by type I and type III interferon (2–4). How Mx1 proteins inhibit viral replication is still largely undetermined. It has been shown that mouse Mx1 can suppress primary transcription of influenza A virus (IAV) genes in the nucleus (5). Furthermore, we previously reported that murine Mx1 can interact with the polymerase basic 2 (PB2) protein and nucleoprotein (NP) in IAV ribonucleoproteins (vRNPs) and disturb the PB2-NP interaction (6). Human MxA, the orthologue of mouse Mx1, can also interact with IAV NP (7). Moreover, NP has been shown to be a determinant of the sensitivity of IAVs for Mx1 and MxA (8, 9). Based on these studies, and on the observation that human MxA—like dynamins—can form ring-like structures (10–14), we hypothesized that the interaction with IAV PB2 and NP might be mediated by a ring structure comprised of oligomerized Mx1, which then actively disrupts the PB2-NP interaction (6). Indirect support for this hypothesis was obtained from the observation that an Mx1 construct that was only active in the presence of an artificial small compound drug could disrupt preexisting IAV vRNPs (15).
The GTPase activity of Mx1 and MxA is required for the suppression of IAV replication. Presumably, the GTPase function, combined with the 2 hinges that flank the central bundle signaling element that separates the globular head domain from the extended helical stalk domain, allows Mx proteins to function as molecular machines that exert a kind of “power stroke.” This mechanochemical transition might generate latitudinal shear forces between neighboring Mx ring structures that destroy the functional vRNP structure (16). In addition to their antiviral effect against IAV, Mx proteins can also restrict replication of Thogoto virus (THOV, a member of the Orthomyxoviridae family). Mouse Mx1, which is only active in the cell nucleus, inhibits THOV multiplication (17). It has also been shown that human MxA can interact with the NP molecules of the THOV vRNPs. This interaction prevents THOV vRNPs from entering the nucleus (18, 19).
The Orthomyxoviridae family currently comprises the following seven genera: Influenza A, B, C and D; Thogotovirus; Quaranjavirus; and Isavirus (20–22). Influenza A and B viruses are important human respiratory pathogens. THOV is a tick-borne virus that has small rodents as natural hosts and very rarely causes zoonotic infection (23, 24). When people become infected with IAV, the first cells that are targeted are the airway epithelial cells. After binding, endocytosis, and membrane fusion, the viral vRNPs are released into the cytoplasm. These then enter the nucleus, where transcription and replication will take place (reviewed in te Velthuis and Fodor [25]). The incoming vRNPs first direct the synthesis of viral mRNA (primary transcription), which is transported to the cytosol and translated. Newly produced PB1, PB2, polymerase acidic protein (PA), and NP migrate to the nucleus to initiate replication of the viral genome and boost transcription. The resulting progeny viral RNA molecules form vRNPs and leave the nucleus, ready for packaging and budding (26). Although THOV has not been studied as elaborately as IAV, it has been shown that both viruses are structurally and genetically similar (27–33). Several studies have also pointed out that their replication cycles are comparable (17–19, 34–40). When a mouse becomes infected with THOV, the virus replicates and spreads rapidly to different sites in the mouse body to eventually kill the mouse (41). A similar pathogenesis in mice following infection with the related Dhori virus has been reported; the virus could be detected in multiple organs, such as the brain, lungs, thymus, spleen, adrenal glands, and liver (42). However, the main target organ of THOV and Dhori virus is the liver, where these viruses can replicate to very high titers and cause severe coagulative zonal necrosis, leading to the rapid death of the infected mouse (17, 42).
Mx proteins exert their antiviral activity in a cell-autonomous way. Haller and coworkers reported that athymic (nude) mice, which carry a functional Mx1 gene, survived intracerebral infection with a neurotropic IAV strain, demonstrating that Mx1-positive mice do not require a functional T cell system to survive the infection (43). Later, the same lab reported that in vivo resistance to a pneumotropic, neurotropic, or hepatotropic strain of IAV was largely independent of whether macrophages carried a functional Mx1 gene or not (44). These studies showed that Mx1 expression in hematopoietic cells does not play a major role in resistance against IAV infection. It is also important to note that almost all mouse genetic studies on the contribution of interferon-induced gene products to antiviral defense in the immune cell compartment have been carried out in laboratory mouse strains that lack a functional Mx1 gene (45).
The aim of the present study is to examine the role of Mx1 expression in immune cells for the antiviral host defense. We show that in bone marrow chimeric mice the protection by Mx1 against IAV infection depends solely on the genotype of the stromal (nonhematopoietic) cells, as previously demonstrated by Haller et al. (43). In striking contrast, challenge infection with THOV showed that Mx1 expression in bone marrow-derived cells is sufficient to markedly reduce virus replication and dissemination and delay morbidity in mice.
RESULTS
Resistance to influenza A virus infection primarily depends on the Mx1 genotype of the recipient.
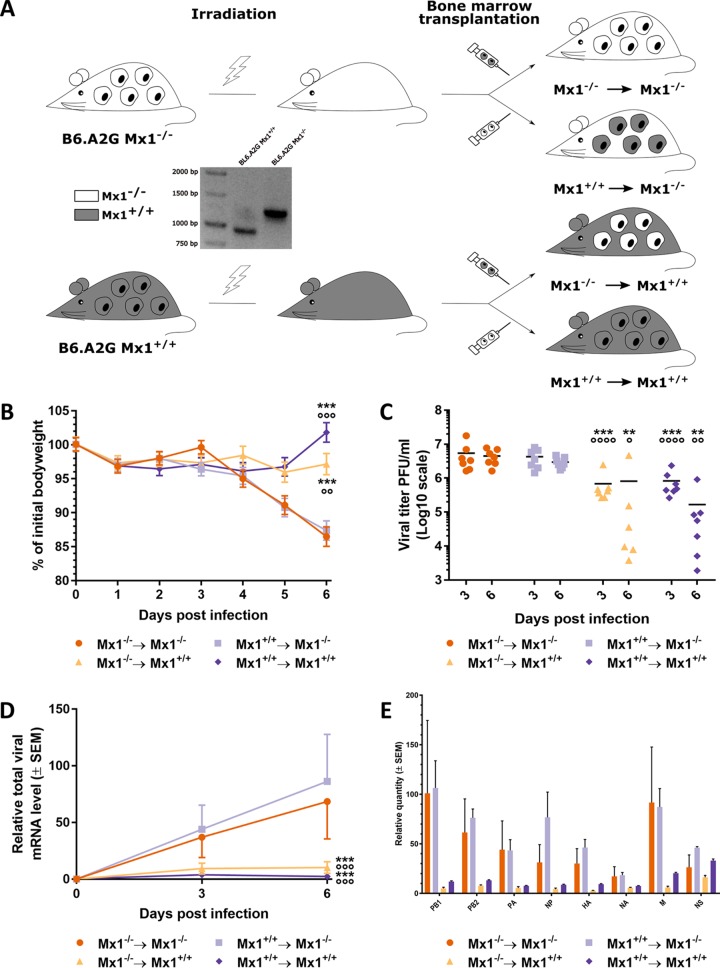
Most immune cells originate from multipotent hematopoietic stem cells in the bone marrow. To address the possible role of Mx1 as a virus restriction factor in this compartment, we generated all four possible bone marrow chimeric mice between B6.A2G Mx1−/− (Mx1−/−) and B6.A2G Mx1+/+ (Mx1+/+) mice (Fig. 1A). Eight weeks after bone marrow transfer, the chimeric mice were infected with 10 lethal dose 50% (LD50) of maPR8 virus, and body weight was monitored for 6 days postinfection (dpi). B6.A2G Mx1−/− recipient mice displayed significantly more bodyweight loss than Mx1+/+ recipient mice, regardless of the donor genotype (Fig. 1B). In addition, infection with maPR8 virus of mice with a functional Mx1 gene in the stromal cells did not result in body weight loss regardless of the donor genotype (Fig. 1B). Lung virus loads were significantly lower in the Mx1+/+ recipients than in Mx1−/− recipients on day 3 and 6 after infection. We observed no significant difference in viral loads between Mx1−/− mice that had been reconstituted with bone marrow from either donor, or between the Mx1+/+ recipients that were reconstituted with Mx1−/− or Mx1+/+ donor bone marrow (Fig. 1C). It has been reported that in cells that stably express Mx1, primary transcription of IAV genes is reduced, and this reduction is more pronounced for the larger genes encoding the polymerase subunits compared with the shorter viral transcripts (5). To ascertain that such a differential effect might also be observed in vivo, we used real-time quantitative PCR (RT-qPCR) to quantify the individual viral mRNA levels in the mouse lung on day 3 and 6 after infection. Three and 6 days after infection, the viral mRNA levels were much lower in the lungs of Mx1+/+ mice compared to those in the lungs of Mx1−/− recipient mice (Fig. 1D). In contrast to what was previously reported by Pavlovic et al. (5), we noticed that the inhibiting effect of Mx1 was equally strong for the shorter and longer viral RNA segments (Fig. 1E). Together, these data show that the Mx1 genotype of the recipient rather than that of the donor determines the outcome of maPR8 virus infection, both in terms of controlling disease and viral replication.
FIG 1.
Mx1-mediated resistance to IAV infection primarily depends on the genotype of the recipient bone marrow chimeric mice. (A) Schematic overview of the generation of the bone marrow chimeras. The PCR-based genotyping of the donor and acceptor mice is also depicted. (B) Mice (n = 14 per group) were infected intranasally with 10 LD50 of maPR8 virus, and the body weight change over time after infection was monitored. Data points represent the average of 14 mice from 0 dpi until 3 dpi and the average of 7 mice from 4 dpi until 6 dpi. Error bars represent the standard error of the mean. Asterisks indicate the significant difference between the Mx1−/− → Mx1−/− group and all other groups over time. ***, P < 0.001. Circles indicate the significant difference with the Mx1+/+ → Mx1−/− group over time. ○○, P < 0.01; ○○○, P < 0.001. (C) Mice were sacrificed on 3 and 6 days postinfection, and lung viral loads were determined. Each data point represents the lung viral titer of a single animal. Asterisks indicate the significant difference between the Mx1−/− → Mx1−/− group and all other groups. **, P < 0.01; ***, P < 0.001. Circles indicate the significant difference between the Mx1+/+ → Mx1−/− group and all other groups. ○, P < 0,05; ○○, P < 0,01; ○○○○, P < 0.0001. (D) Viral mRNA load in lung homogenates from mice sacrificed on day 3 and 6 after infection as determined by real-time quantitative PCR (RT-qPCR). Data points represent the average levels of total viral mRNA relative to household genes. Error bars represent standard error of the mean. Asterisks indicate the significant difference between the Mx1−/− → Mx1−/− group and all other groups. ***, P < 0.001. Circles indicate the significant difference between the Mx1+/+ → Mx1−/− group and all other groups. ○○○, P < 0.001. (E) Viral mRNA load of the eight separate influenza virus genome segments in lung homogenates from the respective bone marrow chimeric mice sacrificed on day 6 after infection as determined by RT-qPCR. Bars represent the average levels of viral mRNA relative to household genes. Error bars represent standard error of the mean. The data are pooled from 2 independently performed experiments (first experiment n = 6 and second experiment n = 8).
Mx1 in hematopoietic cells contributes to the control of Thogoto virus infection.
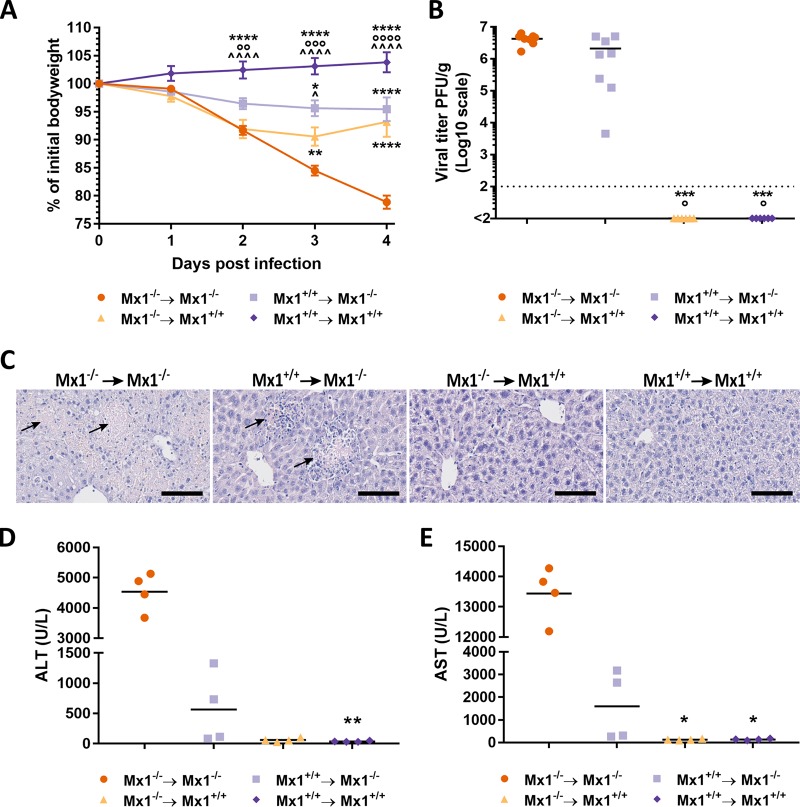
We next addressed the possible contribution of Mx1 in stromal versus hematopoietic cells for control of THOV infection. The rationale for choosing this virus is 3-fold: (i) THOV, like IAV, is a member of the Orthomyxoviridae, (ii) the virus is also sensitive to murine Mx1 (17), and (iii) small rodents are natural hosts of this virus (24). Bone marrow chimeric mice were infected intraperitoneally with 1,000 PFU of THOV (Sicilian SiAr 126 isolate), and body weight and morbidity were monitored daily for 4 days after infection. Mx1+/+ mice that received Mx1+/+ bone marrow showed no signs of morbidity or body weight loss, and Mx1−/− recipients that had been reconstituted with Mx1−/− bone marrow lost significantly more body weight than all other chimeric groups and became moribund by day 4 postinfection (Fig. 2A). Interestingly, Mx1+/+ mice that had received Mx1−/− bone marrow lost significantly more bodyweight than Mx1−/− mice that had received Mx1+/+ bone marrow, indicating a protective role for Mx1 expression in hematopoietic cells in this infection model. Surprisingly, liver viral titers did not reflect these findings. The THOV titers in the liver of the Mx1−/− recipient mice were very high (approximately 106 to 107 PFU/g; the 2-fold lower virus load in the Mx1+/+ recipients did not reach statistical significance) (Fig. 2B). In contrast, THOV virus could not be detected in liver extracts from infected Mx1+/+ recipient mice at 4 dpi, independent of the donor genotype (Fig. 2B). These data suggest that Mx1 expression in hematopoietic cells contributes to the control of THOV-associated morbidity and, to a limited extent, viral replication in the liver of Mx1−/− recipient mice.
FIG 2.
Mx1+/+ hematopoietic cells contribute to protection against THOV-associated pathology. Bone marrow chimeric mice were infected intraperitoneally with 1,000 PFU of THOV on day 0. (A) Body weight change over time after infection. Data points represent the average of eight mice. Error bars represent the standard error of the mean. Statistical analysis was done using a 2-way analysis of variance (ANOVA) with post hoc Tukey’s honestly significant difference (HSD) test. Asterisks indicate the significant difference between the Mx1−/− → Mx1−/− group and all other groups. *, P < 0.05; **, P < 0.01; ****, P < 0.0001. Circles indicate the significant difference between the Mx1+/+ → Mx1−/− group and all other groups. ○○, P < 0.01; ○○○, P < 0.001; ○○○○, P < 0.0001. Caps indicate the significant difference between the Mx1−/− → Mx1+/+ group and all other groups. ^, P < 0.05; ^̂̂̂, P < 0.0001. (B) Viral titers determined by plaque assay in the liver on day 4 after infection. Each data point represents the liver viral titer of a single animal. Statistical analysis was performed using Kruskal-Wallis test with post hoc Dunn’s multiple comparison test. Asterisks indicate the significant difference between the Mx1−/− → Mx1−/− group and all other groups. ***, P < 0.001. Circles indicate the significant difference between the Mx1+/+ → Mx1−/− group and all other groups. °, P < 0.05. Data are pooled from 2 independently performed experiments (first experiment, n = 4; second experiment, n = 4). (C) Histological analysis of liver tissue section (5 μm slides) stained with hematoxylin and eosin after 4 days of infection. Arrows indicate focal zones of liver cell necrosis. Bar, 100 μm. Pictures are representative for n = 8. Serum concentrations of ALT (D) and AST (E) determined on day 4 after infection. Each data point represents the ALT or AST concentration of a single animal (n = 4). Asterisks indicate the significant difference between the Mx1−/− → Mx1−/− group and all other groups. *, P < 0.05; **, P < 0.01; Kruskal-Wallis test with post hoc Dunn’s multiple comparison test.
Mx1+/+ expression in hematopoietic cells reduces THOV-associated liver pathology in Mx1−/− recipients.
THOV infection was previously shown to cause severe liver pathology in Mx1−/− mice, but not in Mx1+/+ mice (17). To examine the possible contribution of donor-derived Mx1 to THOV-associated liver damage, we performed histological analysis on the bone marrow chimeric mice sacrificed on day 4 after THOV infection. Livers isolated from Mx1−/− mice that had been reconstituted with Mx1−/− bone marrow appeared very pale and friable in comparison to the livers from mice in the three other groups, which had a normal brown-red color and firm tissue. Liver sections were prepared, stained with hematoxylin and eosin (H&E), and analyzed microscopically. H&E-stained liver tissue from THOV infected Mx1−/− mice reconstituted with Mx1−/− bone marrow showed lesions with focal to widespread liver cell necrosis (Fig. 2C). Interestingly, liver tissue from Mx1+/+ recipient mice reconstituted with Mx1−/− bone marrow also showed lesions, although these lesions appeared to be in an earlier stage of necrosis compared to those in mice that are Mx1−/− in both the stromal and immune cell compartment. This is manifested as foci in the liver tissue where structure and cell architecture are lost but cell nuclei are still observed. Mx1+/+ recipient’s liver tissue appeared normal and showed no lesions, irrespective of the donor genotype (Fig. 2C). In summary, these results show that Mx1 in cells with a hematopoietic origin can reduce or at least delay liver pathology associated with THOV infection in Mx1−/− recipient mice.
To quantify the degree of liver damage resulting from the THOV infection, alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels were determined in serum of the bone marrow chimeric mice at 4 dpi. The serum levels of both ALT and AST were markedly increased in THOV-infected Mx1−/− mice reconstituted with Mx1−/− bone marrow compared with those in the two Mx1+/+ recipient groups (Fig. 2D and E). Interestingly, THOV infection of irradiated Mx1−/− mice reconstituted with Mx1+/+ bone marrow was associated with intermediate levels of both enzymes. This is in accordance with the histological scoring of the liver sections, further substantiating that Mx1+/+ hematopoietic cells can contribute to protection against THOV infection in mice that lacked Mx1 in the stromal compartment.
Delayed THOV-associated morbidity in Mx1−/− mice grafted with Mx1+/+ bone marrow.
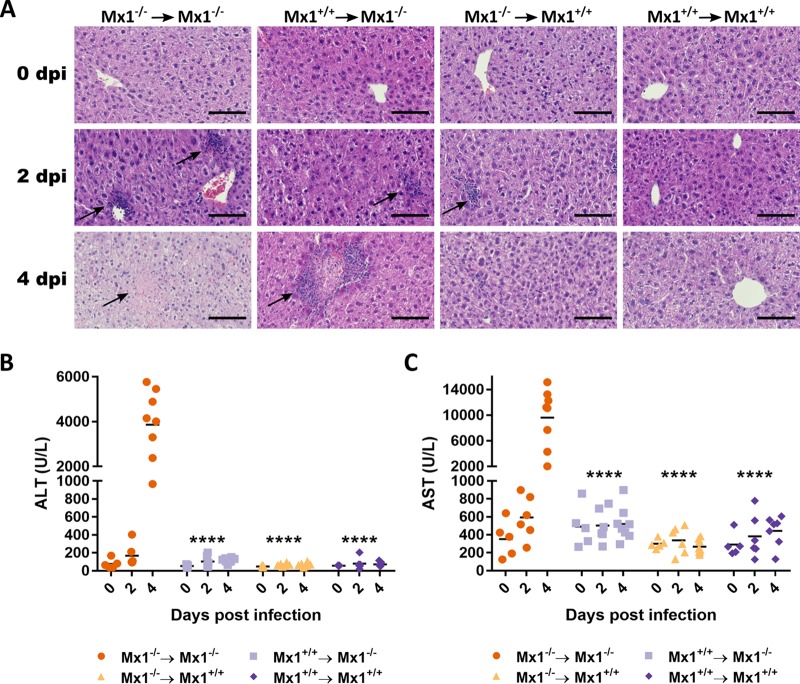
Previous results led to the hypothesis that Mx1 expression in the hematopoietic compartment can delay the course of the THOV infection-associated pathology in Mx1−/− recipient mice. Therefore, we performed additional THOV experiments in the chimeric mice to compare virus replication and pathology at day 2 and 4 after infection. Before infection, all mice had healthy liver tissue (Fig. 3A). Two days after infection, Mx1−/− recipient mice already showed clear zones of cellular influx (Fig. 3A). These zones were markedly larger and more numerous in the Mx1−/− mice reconstituted with Mx1−/− bone marrow than in the Mx1+/+ mice reconstituted with Mx1−/− donor cells. Among the Mx1+/+ recipient mice that had received Mx1−/− bone marrow, a few small zones of cellular influx were observed after 2 days of infection. Four days after infection, Mx1+/+ recipient mice showed no zones of cellular influx or liver cell necrosis. In contrast, all Mx1−/− recipients showed clear zones of cellular influx and/or liver cell necrosis. Livers from mice that lacked Mx1 in both the stromal and immune cell compartment showed clear zones of liver cell necrosis and only a few small zones of cellular influx. Interestingly, livers from Mx1−/− mice that had received Mx1+/+ donor cells showed large zones of cellular influx. In the largest influx zones, a center of necrotic cells could be observed (Fig. 3A, arrows). This indicates that the THOV infection-related pathology is at an earlier state in Mx1−/− mice that received Mx1+/+ bone marrow than that in Mx1−/− mice that received Mx1−/− bone marrow.
FIG 3.
Mx1+/+ immune cells delay THOV-associated liver damage in Mx1−/− recipient mice. Radiation chimeric mice (n = 19 per group) were generated and infected intraperitoneally with 1,000 PFU of THOV. Before infection (n = 5) and on days 2 (n = 6) and 4 (n = 8) after infection, mice were sacrificed and liver and serum samples were prepared. (A) Representative photo micrographs of liver section stained with hematoxylin and eosin. Arrows indicate focal zones of liver cell necrosis. Scale bar = 100 μm. Serum concentrations of ALT (B) and AST (C). Each data point represents the ALT or AST concentration of a single animal. Asterisks indicate the significant difference between the Mx1−/− → Mx1−/− group and all other groups. ****, P < 0.0001. Data are pooled from 2 independently performed experiments (first experiment, n = 9; second experiment, n = 10).
Four days after THOV infection, it was clear that Mx1−/− mice that received Mx1−/− bone marrow showed the highest ALT and AST levels, with averages of approximately 4,000 and 10,000 U/liter, respectively. Mx1+/+ recipients showed only background levels of ALT and AST. In Mx1−/− mice reconstituted with Mx1+/+ bone marrow, intermediate ALT and AST levels were detected in the serum (approximately 100 and 500 U/liter, respectively) (Fig. 3B and C). The differences between the ALT and AST levels between the four groups are most pronounced at 4 dpi. At 2 dpi, the ALT and AST levels of the Mx1−/− mice that received Mx1−/− bone marrow are comparable to the AST and ALT levels of the Mx1−/− mice reconstituted with Mx1+/+ bone marrow at 4 dpi. This shows that the presence of a functional Mx1 gene in hematopoietic cells can delay THOV infection-related liver pathology in mice.
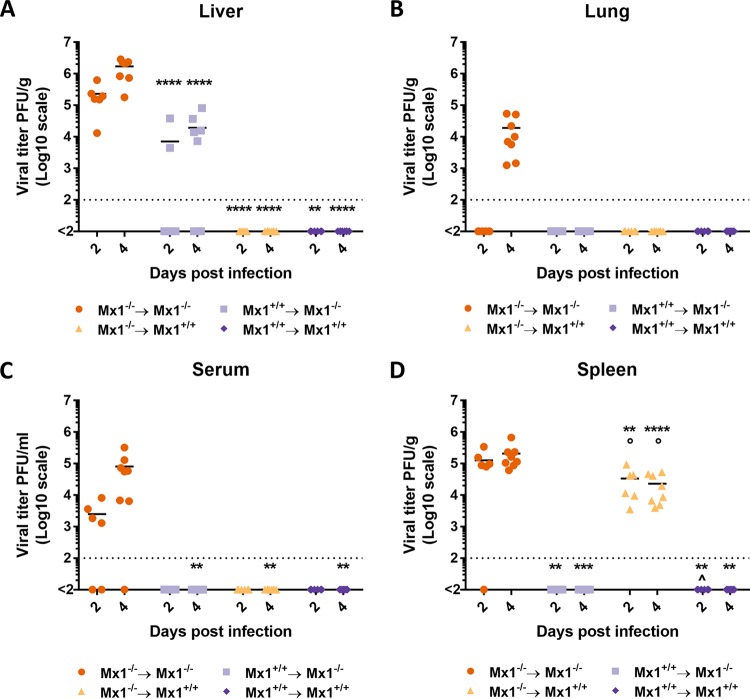
We also assessed the presence of THOV in the spleen, lungs, and serum next to the liver. Similarly to the experiment shown in Fig. 2, liver viral titers at 4 dpi were highest in the Mx1−/− mice that received Mx1−/− bone marrow, 1.5 to 2 logs lower in Mx1−/− mice that received Mx1+/+ bone marrow, and undetectable in both Mx1+/+ recipient groups (Fig. 4A). At 2 dpi, liver viral titers in both Mx1−/− recipient groups are lower compared to the viral titers at 4 dpi. Mx1+/+ recipient mice had undetectable liver viral titers at 2 dpi. THOV was detectable in lung and serum sampled from Mx1−/− mice that received Mx1−/− bone marrow but in none of the other groups (Fig. 4B and C). Strikingly, the spleen viral titers show a remarkable difference compared to the titers in the liver, lungs, and serum. Spleen THOV titers were highest in the Mx1−/− mice that received Mx1−/− bone marrow group at 2 and 4 dpi (Fig. 4D). However, spleen viral titers are below the detection limit in Mx1−/− mice that received Mx1+/+ bone marrow. Surprisingly, Mx1+/+ mice that received Mx1−/− bone marrow show viral titers that are only 4-fold lower than Mx1−/− mice that received Mx1+/+ bone marrow at 2 dpi and 9-fold lower at 4 dpi. Thus, a functional Mx1 gene in the hematopoietic cellular compartment is sufficient to control THOV spread to the lungs, spleen, and serum.
FIG 4.
Mx1 in hematopoietic cells differentially controls THOV replication in different parts of the body. Bone marrow chimeric mice (n = 14 per group) were generated and infected intraperitoneally with 1,000 PFU of THOV. On day 2 (n = 6) and 4 (n = 8) after infection, mice were sacrificed and viral titers in liver (A), lung (B), spleen (C), and serum (D) were determined. Each data point represents the viral titer of a single animal. Asterisks indicate the significant difference between the Mx1−/− → Mx1−/− group and all other groups. **, P < 0,01; ***, P < 0.001; ****, P < 0,0001. Circles indicate the significant difference between the Mx1+/+ → Mx1−/− group and all other groups. ○, P < 0.05. Caps indicate the significant difference between the Mx1−/− → Mx1+/+ group and all other groups. ^, P < 0.05. Data are pooled from 2 independently performed experiments (first experiment, n = 7; second experiment, n = 7).
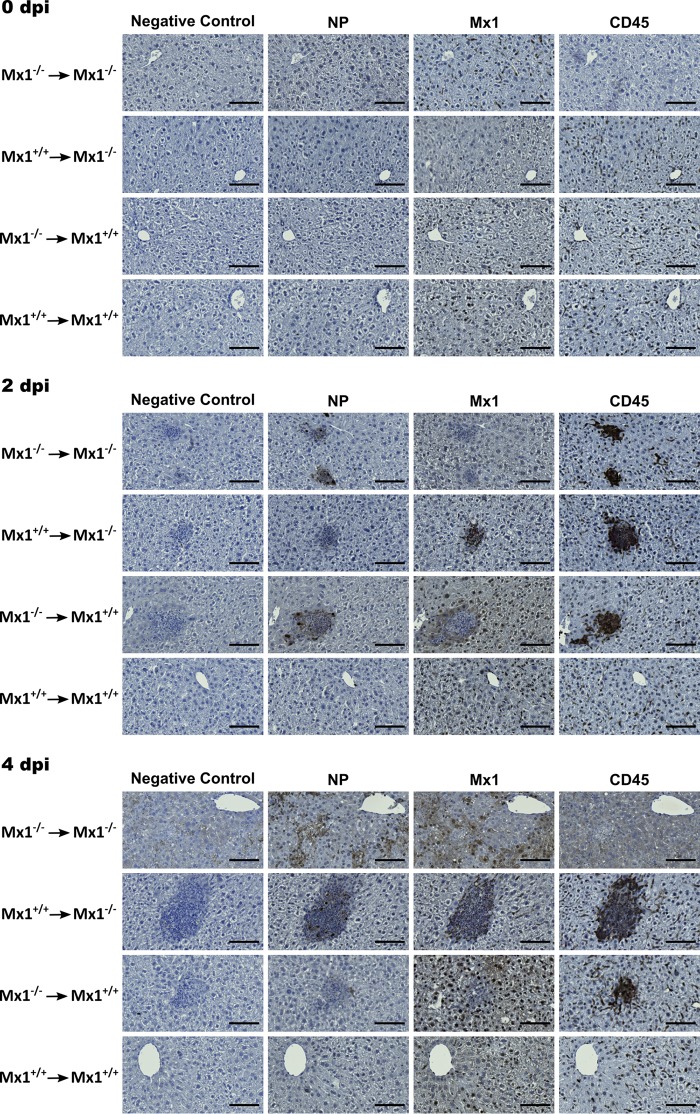
Mx1 expression negatively correlates with THOV NP expression in stromal and bone marrow-derived cells.
The above data show that dissemination of THOV infection depends on the Mx1 genotype of the hematopoietic and/or stromal compartment. To demonstrate the cellular import of THOV infection, liver tissue slides of bone marrow chimeras were stained 2 and 4 dpi with antibodies specific for CD45 (myeloid cell marker), mouse Mx1, or THOV NP. Accumulation of myeloid cells could be detected in all chimeric groups except in Mx1+/+ mice that received Mx1+/+ bone marrow (Fig. 5). In livers of Mx1−/− mice that received Mx1−/− bone marrow, none of the cells stained positive for Mx1. Surprisingly, the only cells that stained positive for THOV NP—cells productively infected by THOV—were cells in the zones of cellular infiltrates. In contrast, cells in the cellular infiltrate zones in livers of Mx1−/− mice that received Mx1+/+ bone marrow stained positive for CD45 and Mx1 but not for THOV NP. In liver tissue of Mx1+/+ mice that received Mx1−/− bone marrow, the opposite was observed. Here, the cells in the cellular infiltrate zone stained positive for CD45 and THOV NP, but not for Mx1. As expected, there were no zones of cellular infiltration in livers of Mx1+/+ mice that received Mx1+/+ bone marrow, and liver cells only stained positive for mouse Mx1. Four days after infection, liver tissue of Mx1−/− mice that received Mx1−/− bone marrow displayed ample liver cell necrosis (Fig. 5). This loss of structure in the liver tissue also influenced the background staining. Except for the large zones of cell infiltration in liver tissue of Mx1−/− mice that received Mx1+/+ bone marrow, staining for CD45, mouse Mx1, and THOV NP at 4 dpi was comparable with that at 2 dpi. Together, these data suggest that cells that make up the zones of cellular infiltration are CD45-positive (CD45+) and that these CD45+ cells first encounter THOV and then infiltrate the peripheral organs and thus disseminate the infection.
FIG 5.
Mx1+/+ hematopoietic cells delay THOV infection-associated liver damage in Mx1−/− recipient mice. Bone marrow chimeric mice (n = 19 per group) were infected intraperitoneally with 1,000 PFU of THOV. Before infection (0 dpi) (n = 5), 2 (n = 6), and 4 (n = 8) days after infection mice were sacrificed, livers were isolated, and prepared for histological analysis by staining tissue slides with hematoxylin and immunostaining with polyclonal antiserum directed against THOV NP and Mx1 or a monoclonal antibody against CD45. Bar, 100 μm.
DISCUSSION
In mice, it is well established that expression of a functional Mx1 protein can protect against a challenge dose of IAV or THOV that otherwise causes severe morbidity and mortality in mice without a functional Mx1 gene. However, whether Mx1 expression is induced in every interferon (IFN)-responsive cell upon infection, and whether Mx1 is needed for their proper functioning, remains an open question. Several studies have shown the importance of an IFN response in cell types involved in the adaptive immune response after IAV infection (46–48). These studies make it tempting to hypothesize that Mx1 can also play a role in protection against viral infection of cell types that are involved in adaptive immunity. Here, we generated Mx1 bone marrow chimeras, allowing us to investigate the possible effect of Mx1 when it is primarily expressed by bone marrow-derived cells or stromal cells. However, some bone marrow-derived cell types, such as Langerhans cells (49) and mesenchymal stromal cells (reviewed in Sugrue et al. [50]), are resistant to lethal total body irradiation. Consequently, these cell types will have the genotype of the bone marrow recipient.
Because of the dominant effect of the presence or lack of Mx1 expression in epithelial cells, the data obtained in this IAV infection model do not answer the question of whether Mx1 can play a role in bone marrow-derived immune cells. Conceivably, the maPR8 virus infection model, characterized by a preferred tropism for epithelial cells, is not best suited for answering the question. It was recently reported that internal genes of highly pathogenic H5N1 viruses can facilitate replication in myeloid cells and lead to severe disease in Mx1-deficient mice (51). It is therefore possible that challenge infections of Mx1 bone marrow chimeras with such influenza viruses could have revealed a more pronounced effect on infection outcome in animals that received Mx1+/+ bone marrow.
Instead of exploring the outcome of infections with a highly pathogenic influenza virus, we turned to THOV challenge infections. Bone marrow chimeras were infected intraperitoneally with a high dose of THOV. Morbidity was somewhat different from that seen in the IAV infection model, in that Mx1−/− mice that received Mx1+/+ bone marrow showed less body weight loss than Mx1+/+ mice that received Mx1−/− bone marrow. Remarkably, liver viral titers at 4 dpi were very high for all of the Mx1−/− recipients and below the detection limit for Mx1+/+ recipients. In order to find an explanation for the seemingly discrepant data, we examined the liver tissue of bone marrow chimeras at microscopic level 4 days after THOV infection. Liver tissue from THOV-infected Mx1−/− recipient mice showed lesions. For the Mx1−/− mice that received Mx1+/+ bone marrow, the cells in these lesions were in an earlier stage of cell necrosis compared to those of Mx1−/− mice that had been grafted with Mx1−/− bone marrow cells, as cell nuclei were still visible. The lesions in these mice were also characterized by a clear cellular influx. As a more objective measure for liver damage, the ALT and AST serum levels following THOV infection were determined. The obtained data reflected the results of the histological analysis of the liver tissue. This suggests that the expression of a functional Mx1 protein in hematopoietically derived cells cannot protect against productive THOV infection in Mx1−/− recipients but can delay disease progression. This was in line with the THOV infection kinetics data. Histological analysis of liver tissue, as well as ALT and AST levels in blood serum, showed that Mx1−/− mice that received Mx1+/+ bone marrow cells have a delayed progression of liver damage compared with Mx1−/− mice that received Mx1−/− bone marrow cells. Another argument for this theory is that the situation in livers of Mx1−/− mice that received Mx1−/− bone marrow at 2 dpi is comparable with that at 4 dpi in livers of Mx1−/− mice that received Mx1+/+ bone marrow. In these two situations, we showed comparable liver morbidity and cellular influx in the liver. Immunohistological analysis made it clear that the cellular influx in the livers is mainly composed of CD45+ immune cells. It was apparent that cells that express a functional Mx1 protein (CD45+ or CD45−) do not express the viral protein NP, which suggests that these cells were not productively infected. Interestingly, in livers of Mx1−/− mice that received Mx1−/− bone marrow, the CD45+ cells appeared to be the first cells that express the THOV NP.
Recently, Kochs et al. postulated that THOV has a tropism for CD11b+ cells with a clear myeloid/macrophage phenotype (double positive for surface markers CD11b and F4/80) in the peritoneum (52). Therefore, it is conceivable that these cells could be partially protected against THOV infection by Mx1 expression. When these cells become infected with THOV, they likely transport the virus to the liver. This is a credible theory, given that Ghosn et al. identified a population of large peritoneal macrophages (LPMs) which seem to have a similar phenotype as the CD11b+ myeloid cells described by Kochs et al. (52, 53). These LPMs can migrate to the omentum, a fat tissue that connects the abdominal organs, upon inflammation (54), which is in agreement with the disappearance of the CD11b+ myeloid cell population from the peritoneal cavity after THOV infection (52). From the omentum, the LPMs can reach the liver of infected mice. There, the virus can infect hepatocytes, unless these hepatocytes express a functional Mx1 protein, in which case viral replication would be suppressed in the hepatocytes. In Mx1−/− mice that received Mx1+/+ bone marrow, we observed high viral titers and necrotic cell lesions in the liver, indicating that THOV can still reach the liver. However, THOV NP expression in the liver was low to nonexistent. It is plausible that myeloid cells are only partially protected against THOV infection, or that the viral inoculum (1,000 PFU) used overcomes the Mx1 restriction in the Mx1+/+ myeloid cells. Nonetheless, the obtained results indicate that Mx1 expression in myeloid cells can delay the progression of THOV infection. Mx1+/+ mice that received Mx1−/− bone marrow showed no detectable liver viral titers and no liver injury. However, these mice showed clear viral titers in the spleen, which could be explained by the high abundance of myeloid cells in this organ. If THOV could reach the liver tissue via THOV-susceptible Mx1−/− myeloid cells, the virus would still be inhibited by the presence of a functional Mx1 protein in the hepatocytes. However, this does not explain the higher weight loss for these mice in comparison with that of the Mx1−/− and Mx1+/+ mice that received Mx1+/+ bone marrow. Conceivably, since THOV can still reach the liver and spleen quite easily in Mx1+/+ mice that received Mx1−/− bone marrow, an inflammatory response will be triggered. This response can cause the production of inflammatory cytokines (55, 56) and is possibly the reason for the more severe weight loss in Mx1+/+ mice that received Mx1−/− bone marrow. Replication in myeloid cells could lead to high type I IFN levels and lead to a cytokine storm, which would explain the severe weight loss in Mx1+/+ mice that received Mx1−/− bone marrow (51).
In conclusion, to confer resistance against Mx1-susceptible viruses that do not have a tropism for myeloid cells, such as influenza A/Puerto Rico/8/34, Mx1 expression is primarily important in the stromal cells. However, for resistance against Mx1-susceptible viruses, like THOV, that can infect myeloid cells and disseminate through these cells, Mx1 expression in bone marrow-derived cells is of major importance.
MATERIALS AND METHODS
Ethics statement.
All animal experiments described in this study were conducted according to the national (Belgian Law 14/08/1986 and 22/12/2003; Belgian Royal Decree 06/04/2010) and European legislation (European Union Directives 2010/63/EU and 86/609/EEC). All experiments on mice and animal protocols were approved by the ethics committee of Ghent University (permit numbers LA1400091 and EC2015-027).
Mice.
Mice were bred in-house under specific pathogen-free (SPF) conditions. Mice were housed in individually ventilated cages in a temperature-controlled environment with 12-h light/dark cycles, with food and water ad libitum. Congenic B6.A2G-Mx1 (Mx1+/+) mice with a functional A2G Mx1 allele were kindly provided by Peter Stäheli (University of Freiburg, Germany). Congenic B6.A2G-Mx1 (Mx1−/−) carrying the defective C57BL/6J Mx1 allele were generated in our laboratory by crossing B6.A2G-Mx1 (Mx1+/+) with C57BL/6J (Mx1−/−) mice, followed by subsequent crossing of the heterozygous offspring. Mouse genomic DNA was isolated from tail biopsies following digestion at 55°C in buffer containing 50 mM Tris-HCl (pH 8.0), 10 mM EDTA, 100 mM NaCl, 0.1% SDS, and 1 mg/ml proteinase K. A PCR was performed using the following primers: 5′-GGAGCTCACCTCCCACATCT-3′, 5′-AGCATGGCTGTGTCACAAGCA-3′, and 5′-CGAAGGCAGTTTGGACCATCT-3′. PCR consisted of a 1-min denaturation step at 94°C, a 1-min annealing step at 61°C, and a 1-min polymerization step at 72°C (40 cycles). The resulting PCR products were visualized by agarose gel electrophoresis.
Bone marrow chimera mice.
Starting 1 week before and until 3 weeks after irradiation, mice were given water containing 0.2% neomycin ad libitum. Mice were subjected to lethal total body irradiation (10 Gray) with an X-Rad 320 biological irradiator (Precision X-Ray [PXi]; North Branford, CT, USA), and 24 h later they were reconstituted with syngeneic or allogeneic bone marrow cells (8 × 106 to 10 × 106) that were harvested from femurs of age-matched mice. Experimental transfers were as follows: B6.A2G Mx1−/− donors into B6.A2G Mx1−/− recipients (Mx1−/− → Mx1−/−), B6.A2G Mx1−/− donors into B6.A2G Mx1+/+ recipients (Mx1−/− → Mx1+/+), B6.A2G Mx1+/+ donors into B6.A2G Mx1+/+ recipients (Mx1+/+ → Mx1+/+), and B6.A2G Mx1+/+ donors into B6.A2G Mx1−/− recipients (Mx1+/+ → Mx1−/−). Animals were allowed to recover and reconstitute their hematopoietic cellular compartment for 8 weeks. Only healthy mice without obvious signs of graft-versus-host disease were used in experiments.
Virus challenge.
Mice were challenged with 10 lethal dose 50% (LD50) (approximately 170 PFU) of mouse adapted (ma) influenza A/Puerto Rico/8/34 (PR8) (H1N1) or with 103 PFU of THOV SiAr 126 (17). The challenge dose was administered intranasally in a volume of 50 μl (maPR8) or intraperitoneally in a volume of 100 μl (THOV) to mice that were anesthetized with a mixture of ketamine (10 mg/kg) and xylazine (60 mg/kg). Morbidity was monitored for 6 (maPR8) or 4 (THOV) days postinfection. Mice that had lost 25% or more of their body weight were euthanized by cervical dislocation.
Determination of influenza lung virus titers.
Mice were sacrificed at different time points after infection by intraperitoneal injection of pentobarbital (125 μg/g). The mouse lungs were removed aseptically, and the left lobe was snap frozen in liquid nitrogen. Lung extracts were prepared by homogenizing the lungs in phosphate-buffered saline (PBS) using metal beads. Cell debris was cleared by centrifugation for 10 min at 400 × g and 4°C. Cleared lung extracts were stored at −80°C. Influenza virus titers were determined in triplicate by titration on MDCK cells. Briefly, MDCK monolayers were infected for 1 h with 500 μl of serial 1:10 dilutions of the lung homogenates in a 12-well plate in serum-free Dulbecco’s modified Eagle medium (DMEM) medium supplemented with penicillin and streptomycin. Following inoculation, the supernatant was replaced by medium containing 2 μg/ml trypsin and 0.6% Avicel RC-851 (FMC Biopolymers). Two days after infection, the cells were fixed with 4% paraformaldehyde, and permeabilized with PBS containing 0.2% Triton X-100. Plaques were stained using a mouse monoclonal antibody against the ectodomain of the influenza M2 protein, and a horseradish peroxidase (HRP)-conjugated anti-mouse IgG antibody (sheep anti-mouse IgG HRP; GE Healthcare, UK). Plaques were then visualized using TrueBlue peroxidase substrate (Seracare, Gaithersburg, MD).
Determination of Thogoto liver virus titers.
Mice were killed by cervical dislocation, the liver was removed aseptically, and one of the lobes was used for histochemistry. Liver extracts were made by homogenizing the livers in PBS using metal beads. Cell debris was cleared by centrifugation for 10 min at 400 × g and 4°C. Cleared liver extracts were stored at −80°C before use. THOV titers were determined in triplicate by titration on Vero cells. Monolayers of Vero cells were infected for 1 h with 1 ml of serial 1:10 dilutions of the liver homogenates in a 6-well plate in DMEM medium supplemented with 2% fetal calf serum and 20 mM HEPES (pH 7.3). Following inoculation, the supernatant was replaced by medium containing 0.6% Avicel RC-951 (FMC Biopolymers). Four days after infection, the cells were fixed with 4% paraformaldehyde. The cell monolayers were stained with a crystal violet solution (1% crystal violet plus 1% methanol plus 20% ethanol) for approximately 15 min at room temperature. The crystal violet solution was then removed, and the wells were washed with water to reveal the plaques.
Histopathological examination of livers.
Livers of bone marrow chimeric mice were excised at 0, 2, or 4 dpi. After fixation in 4% paraformaldehyde (PFA) and embedding in paraffin, livers were sectioned at 5 μm. Sections were used for hematoxylin and eosin staining and immunohistochemical analysis. After incubation with primary and secondary biotin-conjugated antibodies, immunoreactivity was revealed using the ABC-HRP Kit (Vector Laboratories, Burlingame, CA) and the sections were counterstained with hematoxylin. Images were obtained with an Axio Scan.Z1 slide scanner (Zeiss, Oberkochen, Germany) and analyzed with Zen Lite software (Zeiss, Oberkochen, Germany).
ALT/AST assay.
Blood was taken by retroorbital bleeding after sedation of the mice with pentobarbital (125 μg/g). To prepare mouse serum, the blood samples were allowed to clot overnight at 4°C. The next day, the clot was removed and samples were centrifuged at 14,000 rpm for 3 min. Serum samples were stored at −20°C before use. Levels of aspartate aminotransferase (AST) and alanine aminotransferase (ALT) were measured using a Hitachi kit and apparatus in the Clinical Biology Laboratory of Ghent University Hospital.
Real-time quantitative PCR.
Mice of each group were sacrificed just prior to and on day 3 and 6 after IAV infection by intraperitoneal injection of pentobarbital (125 μg/g). The mouse lungs were removed aseptically, and the left lobe was snap frozen in liquid nitrogen. Lung extracts were made by homogenizing the lungs in PBS using metal beads. Cell debris were cleared by centrifugation for 10 min at 400 × g and 4°C. Cleared lung extracts were stored at −80°C until use. RNA was isolated with the High Pure RNA isolation kit (product no. 11828665001, Roche) as indicated by the manufacturer. Total mRNA was converted to cDNA by reverse transcription-PCR (RT-PCR) using an oligo(dT) reaction (Transcriptor First Strand cDNA synthesis kit, catalog no. 04897030001; Roche). cDNA (10 ng) was used for each quantitative PCR (qPCR) reaction, and triplicate reactions were set up in 384-well plates. qPCR reactions based on SYBR green detection were performed using a LightCycler (Roche). qPCR-data were analyzed using the qbase+ software packet (Biogazelle, Zwijnaarde, Belgium).
The primers used in this study are as follows. M1/2 forward: 5′-GGGAAGAACACCGATCTTGA-3′; M1/2 reverse: 5′-CGGTGAGCGTGAACACAAAT-3′; NA forward: 5′-CATCTCTTTGTCCCATCCGT-3′; NA reverse: 5′-GTCCTGCATTCCAAGTGAGA-3′; HA forward: 5′-GAGGAGCTGAGGGAGCAAT-3′; HA reverse: 5′-GCCGTTACTCCGTTTGTGTT-3′; PB1 forward: 5′-CCTCCTTACAGCCATGGGA-3′; PB1 reverse: 5′-GTGCTCCAGTTTCGGTGTTT-3′; PB2 forward: 5′-GGATCAGACCGAGTGATGGT-3′; PB2 reverse: 5′-CCATGCTTTAGCCTTTCGACT-3′; PA forward: 5′-CATCAATGAGCAAGGCGAGT-3′; PA reverse: 5′-GCCCCTGTAGTGTTGCAAAT-3′; NP forward: 5′-CAGCCTAATCAGACCAAATG-3′; NP reverse: 5′-TACCTGCTTCTCAGTTCAAG-3′; NS1 forward: 5′-TTCACCATTGCCTTCTCTTC-3′; NS1 reverse: 5′-CCCATTCTCATTACTGCTTC-3′; HPRT1 forward: 5′-AGTGTTGGATACAGGCCAGAC-3′; HPRT1 reverse: 5′-CGTGATTCAAATCCCTGAAGT-3′; UBC forward: 5′-AGGTCAAACAGGAAGACAGACGTA-3′; UBC reverse: 5′-TCACACCCAAGAACAAGCACA-3′; GAPDH forward: 5′-TGAAGCAGGCATCTGAGGG-3′; GAPDH reverse: 5′-CGAAGGTGGAAGAGTGGGAG-3′; TBP forward: 5′-TCTACCGTGAATCTTGGCTGTAAA-3′; TBP reverse: 5′-TTCTCATGATGACTGCAGCAAA-3′; RPL13A forward: 5′-CCTGCTGCTCTCAAGGTT-3′; RPL13A reverse: 5′-TGGTTGTCACTGCCTGGTACTT-3′; actin forward: 5′-GCTTCTAGGCGGACTGTTACTGA-3′; actin reverse: 5′-GCCATGCCAATGTTGTCTCTTAT-3′.
Antibodies.
A polyclonal antiserum against mouse Mx1 was generated by immunizing New Zealand White rabbits with a synthetic, high-performance liquid chromatography-purified peptide CKKFLKRRLLRLDEARQKLAKFSD (C terminus of the Mx1 protein) and purified as described (6). M2e-specific monoclonal antibody was produced in our laboratory. Briefly, hybridomas that produce M2e-specific monoclonal antibodies were isolated as described (57). After subcloning, these hybridoma cultures were scaled up, and monoclonal antibodies were purified from the culture supernatant with a protein A column (GE Healthcare). Polyclonal anti-Thogoto virus NP antibody (antiserum, rabbit), was generated in the laboratory of Georg Kochs (University of Freiburg, Germany) (32). Polyclonal anti-CD45 antibody (rabbit) was obtained from Abcam (ab10558). Biotinylated anti-rabbit antibody (goat) was obtained from Vector Laboratories (catalog no. BA-1000).
Statistical analysis.
The obtained data were analyzed using GraphPad Prism 7 or Genstat software. Methods used in Genstat are described below. Statistical tests were performed in GraphPad Prism 7 software and are mentioned in the figure legends. Relative body weight data were analyzed as repeated measurements using the residual maximum likelihood (REML) approach implemented in Genstat v19 (58). Briefly, a linear mixed model with replicate, genotype, time, and genotype × time interaction as fixed terms and subject time used as residual term was fitted to the data. Times of measurement were set at equal intervals, and an autoregressive correlation structure of order 1 with equal variances (i.e., homogeneity across time) was selected as best model fit in all cases, based on the Aikake information coefficient. Significances of the fixed terms and significances of changes in differences between genotype effects over time were assessed by an F test. Viral titers were analyzed with a hierarchical generalized linear mixed model (HGLMM; fixed model: Poisson distribution, log link; random model: gamma distribution, log link) implemented in Genstat v19 (58). Titers below the detection limit have been imputed with values generated as a random sample from a skewed left-tailed beta distribution (1, 5). Fixed terms include GENOTYPE, DPI, TISSUE, and their 2-way and 3-way interaction, while REPLICATE was set as a random term. t statistics were used to assess the significance of tissue-specific genotype effects at 2 and 4 dpi (on the transformed scale). Estimated mean values were obtained as predictions from the HGLMM, formed on the scale of the response variable. RT-qPCR data were compared with a generalized linear mixed model (GLMM) (fixed model: Poisson distribution, log link; random model: gamma distribution, log link) as implemented in Genstat v19 (58) fitted to RT-qPCR expression data of PB1, PB2, PA, NP, HA, NA, M, and NS genes simultaneously. The linear predictor vector of the values can be written as follows: log(μ) = η = X × β + Z × ν, where the matrix X is the design matrix for the fixed terms genotype, time, and genotype × time, β is their vector of regression coefficients, Z is the design matrix for the random term (i.e., gene, replicate, and gene × replicate), and ν is the corresponding vector of random effect having a gamma distribution. The significance of the fixed interaction term genotype × time was assessed by a Wald test. Significance of the regression coefficients were assessed by a t test. Estimated mean values and their standard errors were obtained as predictions from the GLMM, formed on the scale of the response variable. A hierarchical generalized linear mixed model (HGLMM; fixed model: Poisson distribution, log link; random model: gamma distribution, log link) implemented in Genstat v19 (58) has been fitted to the ALT and AST data. Fixed terms include GENOTYPE, DPI, and their interaction, while REPLICATE was set as a random term. t statistics were used to assess the significance of time-specific genotype effects (on the transformed scale). Estimated mean values were obtained as predictions from the HGLMM, formed on the scale of the response variable.
ACKNOWLEDGMENTS
We thank Gnomixx for statistical analysis of the data. We thank Peter Stäheli (University of Freiburg, Germany) for the helpful discussions and for providing us with the B6.A2G Mx1+/+ mouse strain. We thank Anne Hoorens (Ghent University Hospital) for expert advice in liver pathology.
This study was supported by IWT-Vlaanderen (Ph.D. student fellowship to J.S.). The work was funded by Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) grant KO 1579/12-1 to G.K.
REFERENCES
- 1.Verhelst J, Hulpiau P, Saelens X. 2013. Mx proteins: antiviral gatekeepers that restrain the uninvited. Microbiol Mol Biol Rev 77:551–566. doi: 10.1128/MMBR.00024-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horisberger MA, Staeheli P, Haller O. 1983. Interferon induces a unique protein in mouse cells bearing a gene for resistance to influenza virus. Proc Natl Acad Sci U S A 80:1910–1914. doi: 10.1073/pnas.80.7.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Staeheli P, Haller O. 1985. Interferon-induced human protein with homology to protein Mx of influenza virus-resistant mice. Mol Cell Biol 5:2150–2153. doi: 10.1128/mcb.5.8.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holzinger D, Jorns C, Stertz S, Boisson-Dupuis S, Thimme R, Weidmann M, Casanova JL, Haller O, Kochs G. 2007. Induction of MxA gene expression by influenza A virus requires type I or type III interferon signaling. J Virol 81:7776–7785. doi: 10.1128/JVI.00546-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pavlovic J, Haller O, Staeheli P. 1992. Human and mouse Mx proteins inhibit different steps of the influenza virus multiplication cycle. J Virol 66:2564–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verhelst J, Parthoens E, Schepens B, Fiers W, Saelens X. 2012. Interferon-inducible protein Mx1 inhibits influenza virus by interfering with functional viral ribonucleoprotein complex assembly. J Virol 86:13445–13455. doi: 10.1128/JVI.01682-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nigg PE, Pavlovic J. 2015. Oligomerization and GTP-binding requirements of MxA for viral target recognition and antiviral activity against influenza A virus. J Biol Chem 290:29893–29906. doi: 10.1074/jbc.M115.681494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zimmermann P, Manz B, Haller O, Schwemmle M, Kochs G. 2011. The viral nucleoprotein determines Mx sensitivity of influenza A viruses. J Virol 85:8133–8140. doi: 10.1128/JVI.00712-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manz B, Dornfeld D, Gotz V, Zell R, Zimmermann P, Haller O, Kochs G, Schwemmle M. 2013. Pandemic influenza A viruses escape from restriction by human MxA through adaptive mutations in the nucleoprotein. PLoS Pathog 9:e1003279. doi: 10.1371/journal.ppat.1003279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carr JF, Hinshaw JE. 1997. Dynamin assembles into spirals under physiological salt conditions upon the addition of GDP and gamma-phosphate analogues. J Biol Chem 272:28030–28035. doi: 10.1074/jbc.272.44.28030. [DOI] [PubMed] [Google Scholar]
- 11.Daumke O, Gao S, von der Malsburg A, Haller O, Kochs G. 2010. Structure of the MxA stalk elucidates the assembly of ring-like units of an antiviral module. Small GTPases 1:62–64. doi: 10.4161/sgtp.1.1.12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao S, von der Malsburg A, Paeschke S, Behlke J, Haller O, Kochs G, Daumke O. 2010. Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature 465:502–506. doi: 10.1038/nature08972. [DOI] [PubMed] [Google Scholar]
- 13.Hinshaw JE, Schmid SL. 1995. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature 374:190–192. doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- 14.Kochs G, Haener M, Aebi U, Haller O. 2002. Self-assembly of human MxA GTPase into highly ordered dynamin-like oligomers. J Biol Chem 277:14172–14176. doi: 10.1074/jbc.M200244200. [DOI] [PubMed] [Google Scholar]
- 15.Verhelst J, Van Hoecke L, Spitaels J, De Vlieger D, Kolpe A, Saelens X. 2017. Chemical-controlled activation of antiviral myxovirus resistance protein 1. J Biol Chem 292:2226–2236. doi: 10.1074/jbc.M116.748806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, Zhang L, Graf L, Yu B, Liu Y, Kochs G, Zhao Y, Gao S. 2017. Conformational dynamics of dynamin-like MxA revealed by single-molecule FRET. Nat Commun 8:15744. doi: 10.1038/ncomms15744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haller O, Frese M, Rost D, Nuttall PA, Kochs G. 1995. Tick-borne thogoto virus infection in mice is inhibited by the orthomyxovirus resistance gene product Mx1. J Virol 69:2596–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kochs G, Haller O. 1999. GTP-bound human MxA protein interacts with the nucleocapsids of Thogoto virus (Orthomyxoviridae). J Biol Chem 274:4370–4376. doi: 10.1074/jbc.274.7.4370. [DOI] [PubMed] [Google Scholar]
- 19.Kochs G, Haller O. 1999. Interferon-induced human MxA GTPase blocks nuclear import of Thogoto virus nucleocapsids. Proc Natl Acad Sci U S A 96:2082–2086. doi: 10.1073/pnas.96.5.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hause BM, Ducatez M, Collin EA, Ran Z, Liu R, Sheng Z, Armien A, Kaplan B, Chakravarty S, Hoppe AD, Webby RJ, Simonson RR, Li F. 2013. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog 9:e1003176. doi: 10.1371/journal.ppat.1003176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCauley JW, Hongo S, Kaverin NV, Kochs G, Lamb RA, Matrosovich MN. 2012. Family Orthomyxoviridae, p 749–761. In King AM, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy: classification and nomenclature of viruses. Ninth report of the International Committee of Taxonomy of Viruses. Elsevier, New York, NY. [Google Scholar]
- 22.Presti RM, Zhao G, Beatty WL, Mihindukulasuriya KA, da Rosa AP, Popov VL, Tesh RB, Virgin HW, Wang D. 2009. Quaranfil, Johnston Atoll, and Lake Chad viruses are novel members of the family Orthomyxoviridae. J Virol 83:11599–11606. doi: 10.1128/JVI.00677-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore DL, Causey OR, Carey DE, Reddy S, Cooke AR, Akinkugbe FM, David-West TS, Kemp GE. 1975. Arthropod-borne viral infections of man in Nigeria, 1964–1970. Ann Trop Med Parasitol 69:49–64. doi: 10.1080/00034983.1975.11686983. [DOI] [PubMed] [Google Scholar]
- 24.Darwish MA, Hoogstraal H, Omar FM. 1979. A serological survey for Thogoto virus in humans, domestic mammals, and rats in Egypt. J Egypt Public Health Assoc 54:1–8. [PubMed] [Google Scholar]
- 25.te Velthuis AJ, Fodor E. 2016. Influenza virus RNA polymerase: insights into the mechanisms of viral RNA synthesis. Nat Rev Microbiol 14:479–493. doi: 10.1038/nrmicro.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lakdawala SS, Fodor E, Subbarao K. 2016. Moving on out: transport and packaging of influenza viral RNA into virions. Annu Rev Virol 3:411–427. doi: 10.1146/annurev-virology-110615-042345. [DOI] [PubMed] [Google Scholar]
- 27.Fuller FJ, Freedman-Faulstich EZ, Barnes JA. 1987. Complete nucleotide sequence of the tick-borne, orthomyxo-like Dhori/Indian/1313/61 virus nucleoprotein gene. Virology 160:81–87. doi: 10.1016/0042-6822(87)90047-X. [DOI] [PubMed] [Google Scholar]
- 28.Kochs G, Weber F, Gruber S, Delvendahl A, Leitz C, Haller O. 2000. Thogoto virus matrix protein is encoded by a spliced mRNA. J Virol 74:10785–10789. doi: 10.1128/jvi.74.22.10785-10789.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leahy MB, Dessens JT, Weber F, Kochs G, Nuttall PA. 1997. The fourth genus in the Orthomyxoviridae: sequence analyses of two Thogoto virus polymerase proteins and comparison with influenza viruses. Virus Res 50:215–224. doi: 10.1016/S0168-1702(97)00072-5. [DOI] [PubMed] [Google Scholar]
- 30.Weber F, Gruber S, Haller O, Kochs G. 1999. PB2 polymerase subunit of Thogoto virus (Orthomyxoviridae family). Arch Virol 144:1601–1609. doi: 10.1007/s007050050613. [DOI] [PubMed] [Google Scholar]
- 31.Weber F, Haller O, Kochs G. 1996. Nucleoprotein viral RNA and mRNA of Thogoto virus: a novel “cap-stealing” mechanism in tick-borne orthomyxoviruses? J Virol 70:8361–8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hagmaier K, Jennings S, Buse J, Weber F, Kochs G. 2003. Novel gene product of Thogoto virus segment 6 codes for an interferon antagonist. J Virol 77:2747–2752. doi: 10.1128/jvi.77.4.2747-2752.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vogt C, Preuss E, Mayer D, Weber F, Schwemmle M, Kochs G. 2008. The interferon antagonist ML protein of Thogoto virus targets general transcription factor IIB. J Virol 82:11446–11453. doi: 10.1128/JVI.01284-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Portela A, Jones LD, Nuttall P. 1992. Identification of viral structural polypeptides of Thogoto virus (a tick-borne orthomyxo-like virus) and functions associated with the glycoprotein. The J General Virology 73:2823–2830. doi: 10.1099/0022-1317-73-11-2823. [DOI] [PubMed] [Google Scholar]
- 35.Yang M, Feng F, Liu Y, Wang H, Yang Z, Hou W, Liang H. 2016. pH-dependent conformational changes of a Thogoto virus matrix protein reveal mechanisms of viral assembly and uncoating. J Gen Virol 97:2149–2156. doi: 10.1099/jgv.0.000551. [DOI] [PubMed] [Google Scholar]
- 36.Siebler J, Haller O, Kochs G. 1996. Thogoto and Dhori virus replication is blocked by inhibitors of cellular polymerase II activity but does not cause shutoff of host cell protein synthesis. Arch Virol 141:1587–1594. doi: 10.1007/BF01718257. [DOI] [PubMed] [Google Scholar]
- 37.Weber F, Jambrina E, Gonzalez S, Dessens JT, Leahy M, Kochs G, Portela A, Nuttall PA, Haller O, Ortin J, Zurcher T. 1998. In vivo reconstitution of active Thogoto virus polymerase: assays for the compatibility with other orthomyxovirus core proteins and template RNAs. Virus Res 58:13–20. doi: 10.1016/S0168-1702(98)00096-3. [DOI] [PubMed] [Google Scholar]
- 38.Albo C, Martin J, Portela A. 1996. The 5′ ends of Thogoto virus (Orthomyxoviridae) mRNAs are homogeneous in both length and sequence. J Virol 70:9013–9017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guilligay D, Kadlec J, Crepin T, Lunardi T, Bouvier D, Kochs G, Ruigrok RW, Cusack S. 2014. Comparative structural and functional analysis of orthomyxovirus polymerase cap-snatching domains. PLoS One 9:e84973. doi: 10.1371/journal.pone.0084973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wagner E, Engelhardt OG, Weber F, Haller O, Kochs G. 2000. Formation of virus-like particles from cloned cDNAs of Thogoto virus. J Gen Virol 81:2849–2853. doi: 10.1099/0022-1317-81-12-2849. [DOI] [PubMed] [Google Scholar]
- 41.Pulverer JE, Rand U, Lienenklaus S, Kugel D, Zietara N, Kochs G, Naumann R, Weiss S, Staeheli P, Hauser H, Koster M. 2010. Temporal and spatial resolution of type I and III interferon responses in vivo. J Virol 84:8626–8638. doi: 10.1128/JVI.00303-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mateo RI, Xiao SY, Lei H, DA Rosa AP, Tesh RB. 2007. Dhori virus (Orthomyxoviridae: Thogoto virus) infection in mice: a model of the pathogenesis of severe orthomyxovirus infection. Am J Trop Med Hygiene 76:785–790. doi: 10.4269/ajtmh.2007.76.785. [DOI] [PubMed] [Google Scholar]
- 43.Haller O, Lindenmann J. 1974. Athymic (nude) mice express gene for myxovirus resistance. Nature 250:679–680. doi: 10.1038/250679a0. [DOI] [PubMed] [Google Scholar]
- 44.Haller O, Arnheiter H, Lindenmann J. 1979. Natural, genetically determined resistance toward influenza virus in hemopoietic mouse chimeras. Role of mononuclear phagocytes. J Exp Med 150:117–126. doi: 10.1084/jem.150.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Staeheli P, Grob R, Meier E, Sutcliffe JG, Haller O. 1988. Influenza virus-susceptible mice carry Mx genes with a large deletion or a nonsense mutation. Mol Cell Biol 8:4518–4523. doi: 10.1128/mcb.8.10.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wakim LM, Gupta N, Mintern JD, Villadangos JA. 2013. Enhanced survival of lung tissue-resident memory CD8+ T cells during infection with influenza virus due to selective expression of IFITM3. Nat Immunol 14:238–245. doi: 10.1038/ni.2525. [DOI] [PubMed] [Google Scholar]
- 47.Helft J, Manicassamy B, Guermonprez P, Hashimoto D, Silvin A, Agudo J, Brown BD, Schmolke M, Miller JC, Leboeuf M, Murphy KM, Garcia-Sastre A, Merad M. 2012. Cross-presenting CD103+ dendritic cells are protected from influenza virus infection. J Clin Invest 122:4037–4047. doi: 10.1172/JCI60659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moltedo B, Li W, Yount JS, Moran TM. 2011. Unique type I interferon responses determine the functional fate of migratory lung dendritic cells during influenza virus infection. PLoS Pathog 7:e1002345. doi: 10.1371/journal.ppat.1002345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Merad M, Manz MG, Karsunky H, Wagers A, Peters W, Charo I, Weissman IL, Cyster JG, Engleman EG. 2002. Langerhans cells renew in the skin throughout life under steady-state conditions. Nat Immunol 3:1135–1141. doi: 10.1038/ni852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sugrue T, Lowndes NF, Ceredig R. 2013. Mesenchymal stromal cells: radio-resistant members of the bone marrow. Immunol Cell Biol 91:5–11. doi: 10.1038/icb.2012.61. [DOI] [PubMed] [Google Scholar]
- 51.Li H, Bradley KC, Long JS, Frise R, Ashcroft JW, Hartgroves LC, Shelton H, Makris S, Johansson C, Cao B, Barclay WS. 2018. Internal genes of a highly pathogenic H5N1 influenza virus determine high viral replication in myeloid cells and severe outcome of infection in mice. PLoS Pathog 14:e1006821. doi: 10.1371/journal.ppat.1006821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kochs G, Anzaghe M, Kronhart S, Wagner V, Gogesch P, Scheu S, Lienenklaus S, Waibler Z. 2016. In vivo conditions enable IFNAR-independent type I interferon production by peritoneal CD11b+ cells upon Thogoto virus infection. J Virol 90:9330–9337. doi: 10.1128/JVI.00744-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ghosn EE, Cassado AA, Govoni GR, Fukuhara T, Yang Y, Monack DM, Bortoluci KR, Almeida SR, Herzenberg LA, Herzenberg LA. 2010. Two physically, functionally, and developmentally distinct peritoneal macrophage subsets. Proc Natl Acad Sci U S A 107:2568–2573. doi: 10.1073/pnas.0915000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okabe Y, Medzhitov R. 2014. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell 157:832–844. doi: 10.1016/j.cell.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Racanelli V, Rehermann B. 2006. The liver as an immunological organ. Hepatology 43:S54–S62. doi: 10.1002/hep.21060. [DOI] [PubMed] [Google Scholar]
- 56.Tilg H. 2001. Cytokines and liver diseases. Can J Gastroenterol 15:661–668. doi: 10.1155/2001/746736. [DOI] [PubMed] [Google Scholar]
- 57.Cho KJ, Schepens B, Seok JH, Kim S, Roose K, Lee JH, Gallardo R, Van Hamme E, Schymkowitz J, Rousseau F, Fiers W, Saelens X, Kim KH. 2015. Structure of the extracellular domain of matrix protein 2 of influenza A virus in complex with a protective monoclonal antibody. J Virol 89:3700–3711. doi: 10.1128/JVI.02576-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baird D, Murray D, Payne R, Soutar D. 2017. An introduction to GenStat for Windows, 19th ed GenStat, VSN International, Hemel Hempstead, UK. [Google Scholar]