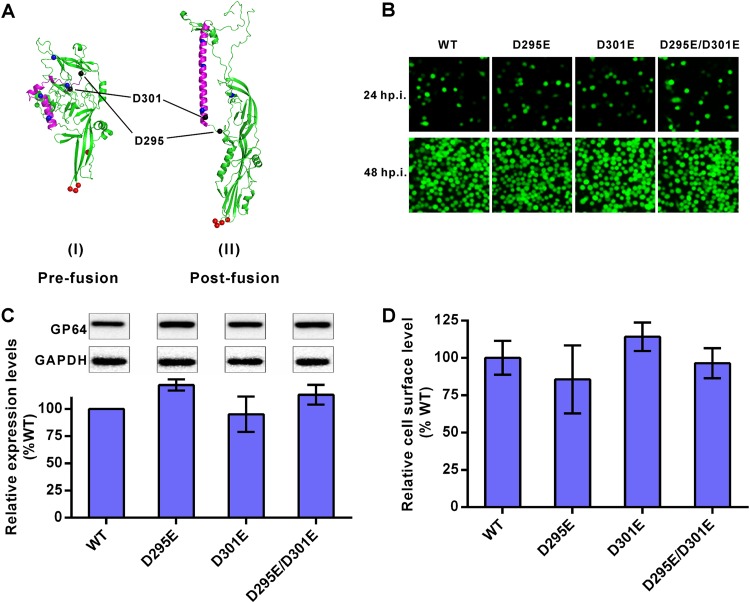
FIG 4.
Construction of recombinant AcMNPV containing mutated GP64. (A) Three-dimensional structures of GP64. Predominant coiled-coil (purple), pH-sensitive histidine (blue dots), fusion loop (red dots), and D295 and D301 (black dots) are indicated. The prefusion structure is a predicted model, and the postfusion structure is based on GP64 data in the Protein Data Bank (PDB code 3DUZ) (48). (B) Infection assay. Sf9 cells were infected by recombinant viruses at an MOI of 1 TCID50 unit/cell and imaged at 24 and 48 h p.i. (C) Expression levels of mutated GP64. Sf9 cells were infected by recombinant viruses at an MOI of 5 TCID50 units/cell and collected for Western blot analysis at 48 h p.i. Expression levels of GP64 were determined by dividing the values for GP64 by those for glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The results were normalized to those of WT GP64. All results are mean values ± SD (n = 3 independent experiments). (D) Analysis of cell surface-displayed mutated GP64. Sf9 cells were infected by recombinant viruses at an MOI of 5 TCID50 units/cell for 48 h and fixed, and the amounts of GP64 protein on cell surfaces were measured via cell surface ELISA. The results were normalized to those of WT GP64. All results are mean values ± SD (n = 3 experimental replicates). For the data shown in panels C and D, statistical analysis was performed using the unpaired t test, and no significant difference was found between the results for the mutants and the WT control (P > 0.05).