FIG 2.
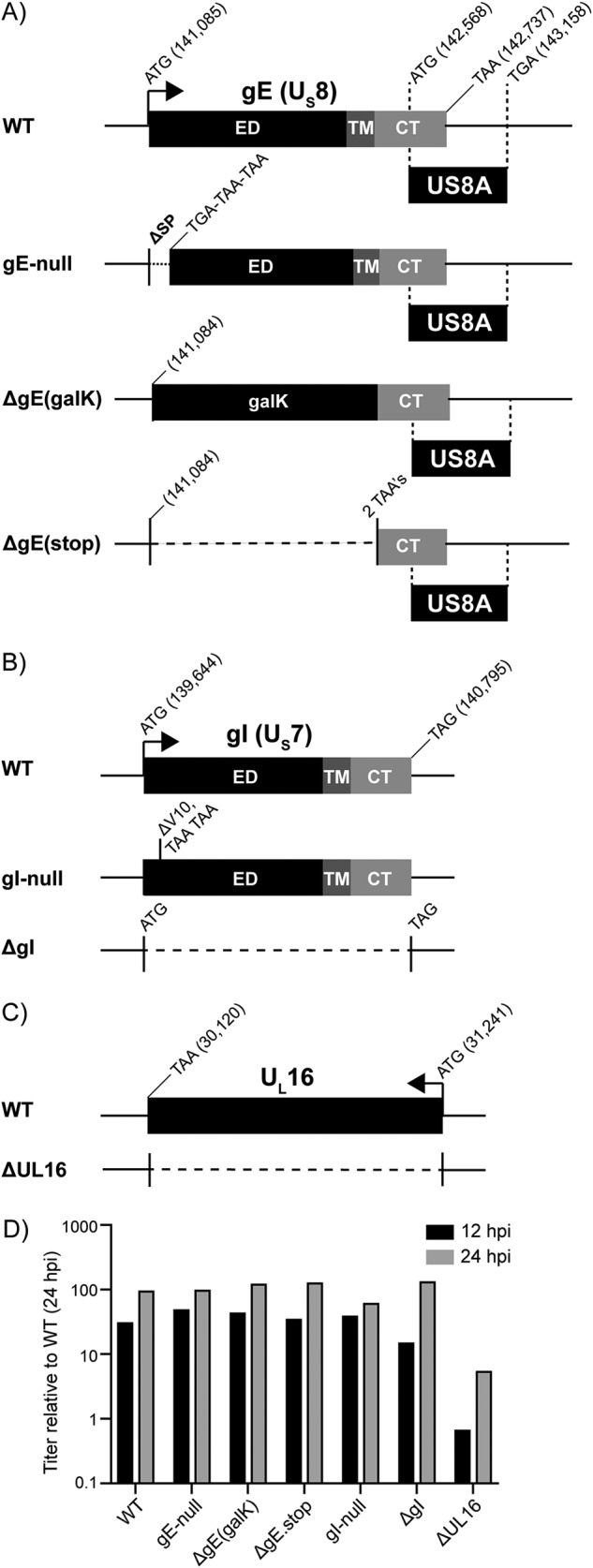
Construction of HSV-1 deletion mutants. (A) An illustration of the US8 (gE) gene and the gE deletion viruses. Sequences encoding the ectodomain (ED), transmembrane domain (TM), and cytoplasmic tail (CT) are indicated. The locations of the start and stop codons within the KOS reference genome are denoted (GenBank accession no. JQ673480.1). Also included is the US8A gene, which overlaps the CT-coding region of US8. The gE-null mutant has three stop codons in place of the signal peptide (SP)-coding sequence. The ΔgE(galK) mutant has a galK cassette in place of the ED-TM-coding sequence. The ΔgE(stop) mutant has two stop codons in place of the ED-TM coding sequence. (B) An illustration of the US7 (gI) gene and the gI deletion viruses. Positions of the ED-, TM-, and CT-coding sequences are indicated. The gI-null mutant has two stop codons in the place of valine codon 10. The ΔgI mutant lacks codons 2 to 383. (C) An illustration of the UL16-coding sequence, which was deleted to make the ΔUL16 mutant. (D) For each deletion mutant, four plates of Vero cells were infected at an MOI of 5. Two plates were harvested at both of the indicated times, and the average titers (cells plus medium) were determined via plaque assays. These were normalized to the wild type at 24 hpi and plotted.
