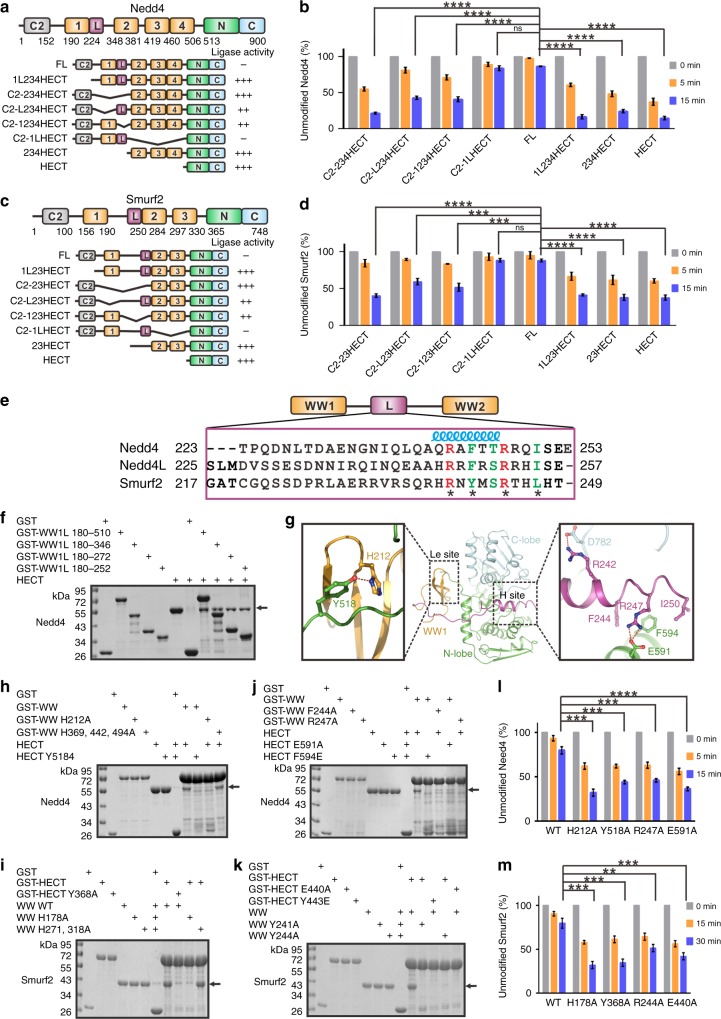
Fig. 6.
C2 and WW1L lock Nedd4 and Smurf2 in an inactive state. a Schematic of Nedd4 domains showing a summary of enzymatic activity derived from the autoubiquitination assay in b. b Statistical results of the autoubiquitination assay of various Trx-tagged Nedd4 fragments in Supplementary Fig. 6b. c Schematic of Smurf2 domains showing a summary of enzymatic activity derived from the autoubiquitination assay in d. d Statistical results of the autoubiquitination assay of various GST-tagged Smurf2 fragments in Supplementary Fig. 6e. e The primary sequence alignment of L in Nedd4, Nedd4L, and Smurf2. The identical residues are colored in red, and the highly conserved residues are colored in green. The residues involved in packing with HECT are marked with asterisks. f GST pull-down assay of GST-tagged Nedd4 WW1L and various truncated fragments with HECT. g Model of Nedd4 1LHECT. h–k GST pull-down assay of Nedd4 GST-WW with Trx-HECT h and j and Smurf2 GST-HECT with Trx-WW i and k. For Nedd4, mutations at the predicted WW1-HECT “Le” site interface (H212AWW1 and Y518AHECT) h and the L-HECT “H” site interface (F244AL, R237AL, E591AHECT, and F594EHECT) j disrupted the interaction between the entire WW region and HECT. For Smurf2, mutations at the predicted WW1-HECT “Le” site interface (H178AWW1 and Y368AHECT) i and the L-HECT “H” site interface (Y241AL, R244AL, E440AHECT, and Y443EHECT) k disrupted the interaction between the entire WW domain region and HECT. l The in vitro autoubiquitination assay of Nedd4 FL mutants (H212AWW1, Y518AHECT, R247AL, or E591AHECT). m The in vitro autoubiquitination assay of Smurf2 FL mutants (H178AWW1, Y368AHECT, R244AL, or E440AHECT). Data are presented as the mean ± SD of triplicate experiments; ns, not significant, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 based on one-way analysis of variance (ANOVA) with Tukey’s multiple comparison test. Source data are provided as a Source Data file