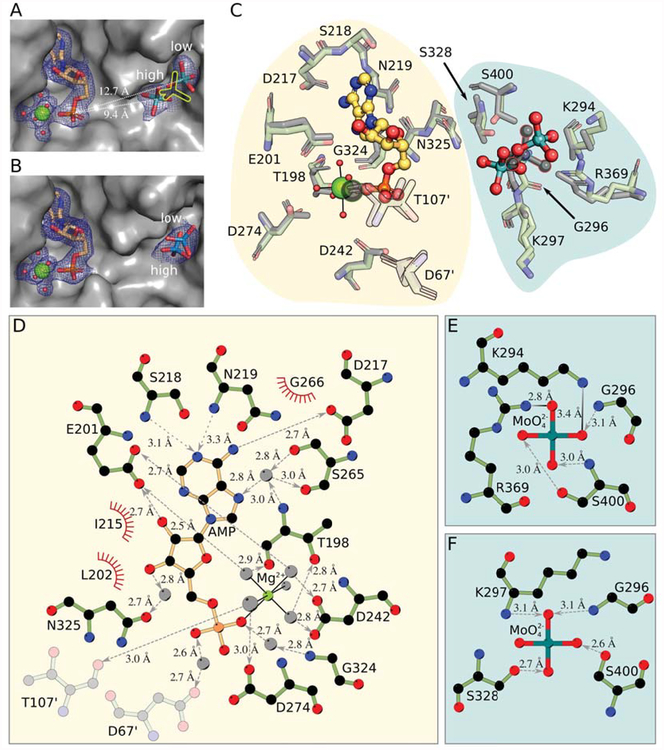
Figure 2: The Cnx1E active site comprising the oxo-anion and AMP binding pockets.
(A) Active site in Cnx1E+Mo with the oxo-anion pocket occupied by two molybdate anions with partial occupancy. The protein is shown as Connolly surface and colored gray. AMP and molybdate are shown as sticks; magnesium ions are shown as big, water molecules are shown as small spheres. Mg2+-AMP and the oxo-anions are surrounded by their corresponding 2Fo-Fc electron density contoured to ı UHSUHVHQWHG E\ D EOXH PHVK 7KH SRVLWLRQ RI WKH PRO\EGDWH DQLRQ IRXQG LQ WKH SUHYLRXVO\ published structure [20] is shown as yellow silhouette. The high- and low-occupancy sites and their distance to the phosphate group of the AMP molecule are indicated. (B) Active site in Cnx1E+W structure with the oxo-anion pocket occupied by two tungstate anions with partial occupancy. The presentation is the same is in panel A. (C) Superposition of the binding pockets of the Cnx1E molybdate co-structure published previously ([20], PDB 5G2S) with the higher resolution Cnx1E+Mo structure. Amino acid residues interacting with Mg2+-AMP or molybdate are shown in simple stick representation whereas the AMP molecule and the molybdate anion are shown in ball-and-stick representation. Magnesium ions are shown as spheres, coordinated water molecules as smaller spheres. The atoms of the previously published structure (2.8 Å, [20]) are colored gray. Residues shown in semi-transparent fashion are located in the other monomer of the physiological dimer. The single previously identified molybdate ion ([20], PDB 5G2S) and the corresponding magnesium ion are highlighted by colored outlines. The background shades are meant to distinguish the two binding pockets of the Cnx1E active site. (D) Schematic representation of the interactions between Cnx1E and Mg2+-AMP. (E) Interactions in the molybdate low-occupancy site, (F) Interactions in the molybdate high-occupancy site. Involved residues and molecules are shown in ball-and-stick representation with disregard for double bonds. The magnesium ion and water molecules are shown as light gray spheres. Interactions within the octahedral Mg2+-water complex are shown as solid black lines. Hydrogen bonds are shown as broken lines. If discernible, arrowheads point to the hydrogen bond acceptor. Salt bridges are represented as solid lines with a color gradient ranging from black to light gray with black indicating the origin of the negative charge in the ionic interaction. Amino acids involved in hydrophobic interactions are represented by red coronas. If not stated otherwise, atoms are colored throughout Fig. 2 as follows: carbon in Cnx1E, green, other carbon, yellow-orange; oxygen, red; nitrogen, blue; phosphorous, orange; magnesium, bright green; molybdenum, cyan; tungsten, teal. Hydrogen atoms are omitted. All distances shown are given in Ångströms.