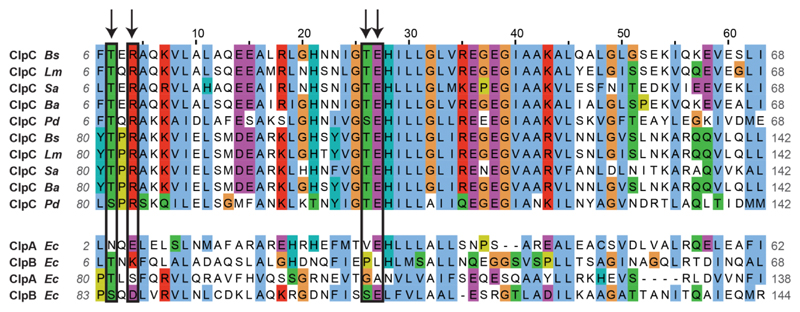
Extended Data Figure 6. Sequence alignment of the pArg-binding site of ClpC from different species and of the homologous regions of other Clp ATPases.
The two symmetrical regions (comprising residues 6–68 and 80–142, approximately) of each protein are aligned. Residues interacting with the pArg molecule (the Glu residue binding to the guanidinium group and the Arg/Thr residues interacting with the phosphate) are circled in black and marked by an arrow. Each of the two pArg-binding sites comprises Glu and Thr from one symmetrical region and Arg and Thr from the other. The alignment shows high conservation of the critical residues of ClpC proteins from different McsB-containing bacteria (B. subtilis, Listeria monocytogenes, Staphylococcus aureus, Bacillus anthracis and Peptoclostridium difficile). Conversely, the residues are not conserved in related Clp proteins (ClpA and ClpB) from McsB-deficient, Gram-negative bacteria.