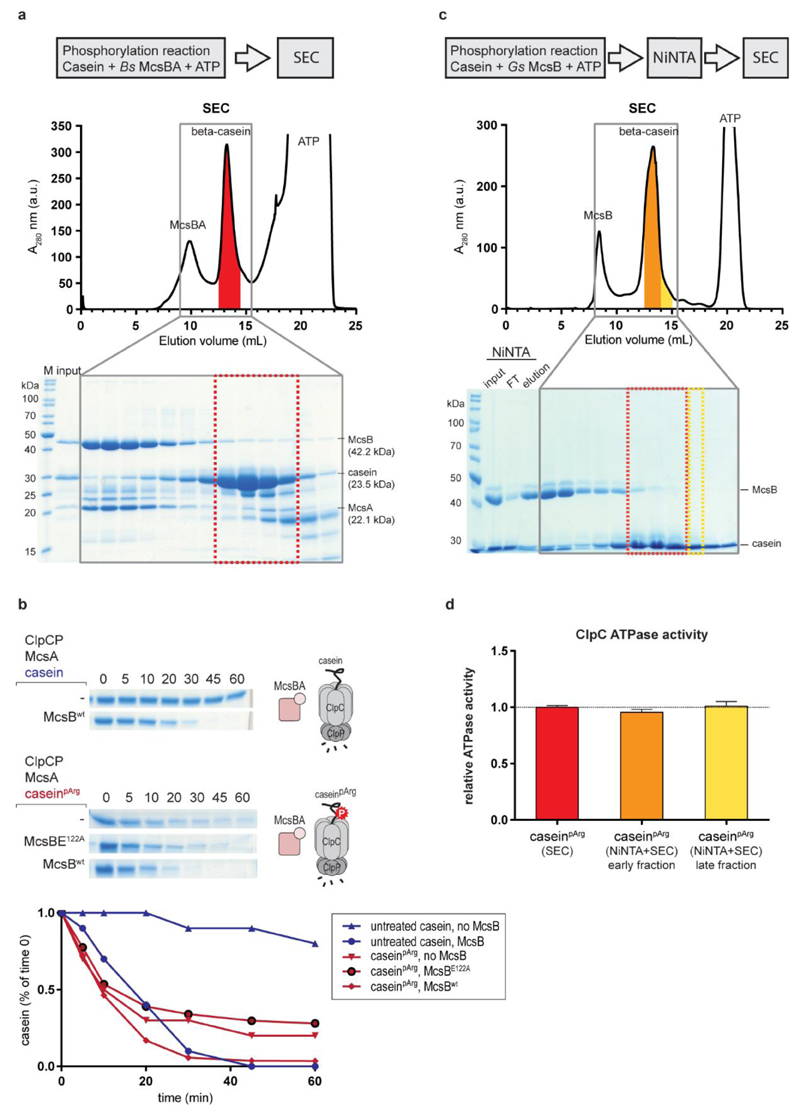
Extended Data Figure 3. Preparation and validation of caseinpArg as a model substrate of ClpCP.
a, Top, size exclusion chromatography (SEC) separation of caseinpArg from the B. subtilis McsBA complex after in vitro phosphorylation. The red markings indicate the fractions that were pooled. Bottom, SDS–PAGE analysis of the fractions indicates that the SEC procedure could separate at least 95% of the McsB protein from the pooled β-casein fractions. b, Evaluating the effect of the McsB contamination on the degradation of caseinpArg. Top, ClpCP in vitro degradation assay towards untreated β-casein. Middle, ClpCP in vitro degradation of caseinpArg, with and without additional McsB kinase. Bottom, addition of either inactive or active McsB did not have an effect on the initial degradation rate of caseinpArg. Of note, the degradation of untreated casein in the presence of McsBA is slightly delayed compared to the degradation of pre-modified caseinpArg. c, An alternative, improved protocol for purifying caseinpArg. After in vitro phosphorylation of β-casein by G. stearothermophilus McsB(6His), a Ni-NTA column was used to capture the tagged kinase. The flow-through fraction was then applied to a SEC column to further separate remaining McsB from β-casein. Two different fractions of the β-casein peak were collected, the later-eluting one (yellow) having a higher degree of purity in relation to the earlier one (orange). d, Activation of the ClpC ATPase activity by the different caseinpArg preparations (2 µM concentration each). Each fraction contains a different (substoichiometric) amount of the McsB contamination. ATPase measurements reveal an almost identical activation of ClpC for all fractions, indicating that the residual amounts of McsB present in the caseinpArg sample do not contribute to ClpC activation. Error bars denote standard deviation of technical triplicates. e, To obtain substrate samples varying in the amount of arginine phosphorylation, casein was pre-incubated with McsBA for increasing times. The YwlE arginine phosphatase was added to the sample phosphorylated most strongly (120 min incubation with McsBA) as a negative control. The resultant caseinpArg samples were subjected to pArg immunoblots using a pArg-specific antibody24 (right, top) and to ClpCP degradation assays (right, bottom).