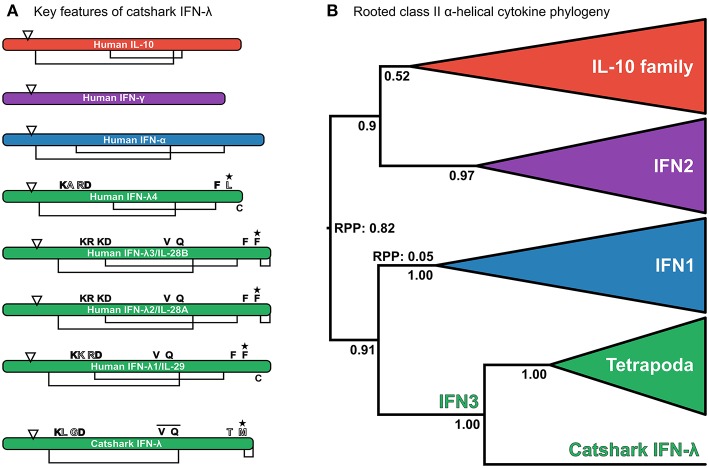
Figure 2.
(A) Analysis of key residues in the catshark IFN-λ sequence compared to a set of other interferons and IL-10. Sequences are represented as cartoon bars, which are relatively scaled according to amino acid sequence length. Arrows denote the end of the signal peptide region, while disulphide bridges are shown as connected regions underneath each cartoon bar, with “C” in the C-terminal region being an unpaired Cys from the characteristic C-terminal disulphide bridge of IFN3s (22). Above the bar the most important residues for IFN-λ3 receptor binding are shown (22). Residues filled in black are conserved, whereas residues filled white are not well conserved, and gray-filled residues involve conserved replacements (e.g., K → R). The bar over the VXXQ motif of catshark IFN-λ indicates that this is not aligned perfectly to human IFN3s, while the star indicates that mutation of this residue abolishes binding in human IFN-λ3 (22). See Figure S1 for full alignment. (B) Relaxed clock (uncorrelated lognormal) rooted class II α-helical cytokine family phylogeny under JTT + I + τ and a Yule speciation prior. The tree is rooted at the best supported root position. Root posterior probabilities (RPP) are shown for branches with a non-negligible probability (i.e., posterior probability <0.05) of being the root. Posterior probabilities are also shown for key nodes, and clades representing individual family members, or the entire IL-10 family have been collapsed to emphasize deep relationships within the family.