SUMMARY
Oomycete RXLR‐dEER effector proteins are rapidly evolving proteins with the selective pressure targeted predominantly at their C‐terminal ends. The majority of RXLR‐dEER proteins have recognizable motifs of 21–30 amino acids in the C‐terminal domain that are named after conserved amino acid residues at fixed positions within the respective motifs. In this article, it is reported that the Phytophthora infestans RXLR‐dEER protein Avr4 contains three W motifs and one Y motif in its C‐terminal domain. Agroinfection assays using constructs encoding modified forms of PiAvr4 have shown that the region containing the W2 motif, in combination with either the W1 or W3 motif, triggers a necrotic response in potato plants carrying the resistance gene R4. By mining the superfamily of avirulence homologues (Avh) deduced from three sequenced Phytophthora genomes, several Avh proteins were identified as homologues of PiAvr4: six in P. infestans, one in P. ramorum and seven in P. sojae. One very close homologue of PiAvr4 was cloned from the sibling species, P. mirabilis. This species is not pathogenic on potato but, similar to PiAvr4, PmirAvh4 triggered a necrotic response on potato clones carrying R4, but not on clones lacking R4. Genes encoding RXLR‐dEER effectors are often located in regions showing genome rearrangements. Alignment of the genomic region harbouring PiAvr4 with syntenic regions in P. sojae and P. ramorum revealed that PiAvr4 is located on a 100‐kb indel block and is surrounded by transposable elements.
INTRODUCTION
Oomycete plant pathogens are responsible for a large number of devastating diseases on many crop plants and ornamentals (Bouwmeester et al., 2009). Like many other plant pathogens, they secrete a range of effector proteins that facilitate the infection of host plants, for example by suppressing defence responses. One class of secreted effectors comprises the RXLR‐dEER proteins that contain two conserved amino acid motifs in the N‐terminal region, RXLR and dEER. These motifs were first found in a number of proteins encoded by oomycete avirulence (Avr) genes that undergo ‘gene‐for‐gene’ interactions with host resistance (R) genes (Govers and Gijzen, 2006; van Poppel et al., 2008; Rehmany et al., 2005). More recent studies have shown that the domain carrying the RXLR and dEER motifs is required for host cell targeting of effector proteins (Dou et al., 2008b; Whisson et al., 2007). Genome mining has revealed that each of the sequenced oomycete genomes contains hundreds of genes encoding RXLR‐dEER effectors (Jiang et al., 2008; Whisson et al., 2007). The majority of RXLR‐dEER genes seem to be derived from a common ancestor and, because of their homology to known Avr genes, they are also referred to as avirulence gene homologues, or Avh genes (Jiang et al., 2008). The three oomycete genomes that have been sequenced and annotated (Phytophthora ramorum, Phytophthora sojae and Phytophthora infestans) show a high degree of conserved synteny (2006a, 2006b). Avh genes, however, are often located on indel blocks and in regions showing genome rearrangements (2008, 2006a). Typically, the Avh genes are flanked by transposon‐like sequences and this may explain the dispersal of these genes throughout the genome (R.H.Y. Jiang and M. C. Zody, Broad Institute, Cambridge, MA, USA, unpublished data).
As postulated in the zig‐zag model (Jones and Dangl, 2006), effectors can evolve to evade host resistance responses. Analyses of the C‐terminal domains of RXLR‐dEER proteins have shown that these effectors are indeed exposed to strong positive selection, leading to fast evolution and diversifying sequences (Jiang et al., 2008; Win et al., 2007). In a recent study, the RXLR‐dEER effector reservoir in two Phytophthora species was analysed and Hidden Markov Model (HMM) searches were used to identify conserved motifs in the C‐terminal region (Jiang et al., 2008). The motifs that were found in over one‐half of all RXLR‐dEER proteins were named W, Y and L after a conserved amino acid residue at a fixed position in the respective motifs. The W (tryptophan), Y (tyrosine) and L (leucine) motifs are 21–30 amino acids in length and can occur in modules in the order W–Y–L. The number of modules and motifs varies in each RXLR‐dEER protein. In P. sojae and P. ramorum, 30% of the RXLR‐dEER proteins possess two to eight W–Y–L modules. Others lack recognizable motifs or have only W motifs, or W and Y motifs. The number of modules correlates with the length of the respective proteins. For example, one RXLR‐dEER protein in P. infestans carries 11 W–Y–L modules on a total length of 989 amino acids (R. H. Y. Jiang, unpublished data). Of the known oomycete Avr proteins, Avr1b from P. sojae has one W and one Y motif, and an additional K motif with several lysine residues (Dou et al., 2008a). Avr3a and IPI‐O from P. infestans each have a single W motif and no Y or L motifs. The Avr proteins ATR1 and ATR13 from Hyaloperonospora parasitica lack W, Y and L motifs, although ATR13 carries several repeats in the C‐terminal region (Allen et al., 2004). As shown by mutational analyses, both the W and Y motifs in Avr1b are involved in governing the avirulence of P. sojae towards soybean plants carrying the resistance gene Rps1b, as well as in suppressing BAX‐mediated programmed cell death (PCD; Dou et al., 2008a). The difference between the virulent and avirulent forms of the P. infestans Avr3a effector is restricted to two amino acids in the C‐terminal region, one of which, at position 103, is located in the W domain (Armstrong et al., 2005). The avirulent form has K80I103, whereas the virulent form has E80M103. The entire 75‐amino‐acid C‐terminal region, including the W motif, is required to elicit the R3a‐dependent hypersensitive response (HR) and to suppress INF1‐triggered PCD; a K80M103 variant, though, is unable to suppress PCD but remains avirulent on R3a plants (Bos et al., 2006). These results suggest a role in avirulence of the sequences flanking the W motif in Avr3a.
Previously, we have identified PiAvr4, a P. infestans avirulence gene that encodes an RXLR‐dEER effector and has a gene‐for‐gene interaction with the potato resistance gene R4 (van Poppel et al., 2008). R4 originates from Solanum demissum and is one of the 11 traditional late blight differentials (Black et al., 1953). All identified late blight R genes encode nucleotide‐binding site–leucine‐rich repeat (NBS‐LRR) proteins and, on the genome, they often co‐localize with other R genes in so‐called R gene clusters. For R4, neither the molecular nature nor its position on the genetic map is known. In this study, we investigated further the recognition of PiAvr4 by R4. We showed that PiAvr4 and its close homologues carry W and Y motifs, and used deletion constructs to investigate which part of the C‐terminus of PiAvr4 and which of the W and Y motifs are required for recognition by potato R4. We also analysed the region in the P. infestans genome that harbours PiAvr4, and demonstrated that, like many Avh genes, PiAvr4 is located at a synteny breakpoint.
RESULTS
Homologues of PiAvr4 in P. infestans and other Phytophthora species
Previously, we have identified several allelic variants of PiAvr4 in different P. infestans isolates and performed genomic Southern blot analysis that suggested the presence of putative PiAvr4 homologues in P. infestans and in sibling species of P. infestans, including Phytophthora phaseoli, Phytophthora andina, Phytophthora mirabilis and Phytophthora ipomoeae (van Poppel et al., 2008). To obtain a PiAvr4 homologue from P. mirabilis, we used PiAvr4‐specific primers for polymerase chain reaction (PCR) amplification and cloned a homologue from strain PIC99111. PmirAvh4 encodes a 290‐amino‐acid protein with high similarity to PiAvr4 (blastp E‐value = 2e‐137; sequence similarity, 89%) and all the characteristics of an RXLR‐dEER protein, including a signal peptide (SP) and an RXLR‐dEER domain. Compared with PiAvr4, PmirAvh4 is three amino acids larger. This is the result of an insertion of six amino acids and a deletion of three amino acids between the RXLR and dEER motifs in PmirAvh4 (Fig. S1, see Supporting Information).
blastn searches of public databases with the PiAvr4 sequence resulted in a single hit to an expressed sequence tag (EST) of P. sojae (AY183415; van Poppel et al., 2008; Qutob et al., 2002). Moreover, 10 RXLR‐dEER proteins, named PsAvh_110, PsAvh_191, PsAvh_192, PsAvh_193, PsAvh_297, PsAvh_342, PiAvh_38, PiAvh_131, PiAvh_190 and PrAvh_40, were identified in a blastp search (amino acid sequence identity >30%) of a pool of Phytophthora Avh, of which PsAvh_110 is the protein corresponding to the previously identified EST. The Avh pool was created by gathering all predicted RXLR‐dEER effectors from three Phytophthora species: 385 in P. sojae, 370 in P. ramorum (Jiang et al., 2008) and 562 in P. infestans (R.H.Y. Jiang and B. Haas, Broad Institute, Cambridge, MA, USA, unpublished data). From subsequent blastp searches using the 10 Avr4 homologues, six additional Avr4 family members were identified (Fig. 1 and Table S1, see Supporting Information). Using the same search criteria, no homologues could be found in the genomes of Phytophthora capsici and H. parasitica.
Figure 1.
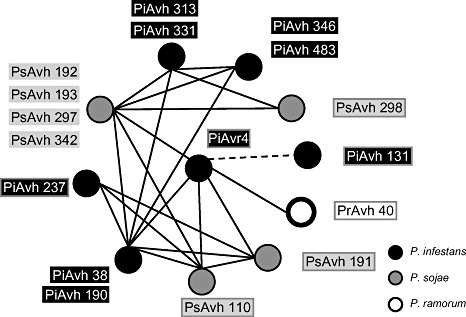
Sequence similarities between PiAvr4 and its homologues. Each protein was used for blastp searches against the entire set of Avh proteins from Phytophthora infestans, P. sojae and P. ramorum. Proteins are represented by circles and labelled next to the circle. Proteins with sequence similarities above 90% are represented by one circle. Different shading of circles and labels is used to distinguish effectors from the different species. A line connecting two proteins represents a blastp hit (E value < 1e‐5). The broken line indicates a similarity that is restricted to the N‐terminus of the two connected proteins.
PiAvr4 in P. infestans is located on a 100‐kb indel block
Comparative analyses of sequenced Phytophthora genomes revealed that the majority of the Avh genes are located on indel blocks that interrupt regions of conserved synteny (2008, 2006a). To determine the genomic context of PiAvr4, we aligned the genomic region surrounding PiAvr4 to the genomic sequences of P. sojae and P. ramorum. PiAvr4 is located on supercontig 11 (size 3761 kb; P. infestans genome assembly version 1.0) at position 359782–360645. Alignment of a 168‐kb region surrounding PiAvr4 with a 45‐kb region on P. sojae scaffold 39 (size 598 kb; genome assembly version 1.1) showed a conserved order and the orientation of 12 gene models with the exception of one inversion (Fig. 2; Table 1). Similarly, a 25‐kb region on P. ramorum scaffold 86 (size 203 kb; genome assembly version 1.1) contains 11 of the gene models in the same order. One gene model encoding an acyltransferase is duplicated in P. sojae and P. infestans, but not in P. ramorum. The most obvious difference between the three genomes is the presence of a 100‐kb indel block in P. infestans that breaks the conserved synteny between the three species. This indel block carries three gene models, one of which is PiAvr4, whereas the other two encode transposons. Apart from these two transposon gene models, the indel block consists almost entirely of transposon‐like sequences. Neither P. sojae nor P. ramorum carries a gene model for a PiAvr4 homologue or any other Avh protein in the syntenic regions on scaffolds 39 and 86, respectively. In P. sojae, the closest PiAvr4 homologue (i.e. PsAvh_110) is located on scaffold 102 and, in P. ramorum (i.e. PrAvh_40), on scaffold 100. This analysis shows that PiAvr4 marks a synteny breakpoint in P. infestans. The fact that PiAvr4 is flanked by many transposon‐like sequences suggests that transposons are involved in the rearrangement of the genomic region carrying PiAvr4.
Figure 2.
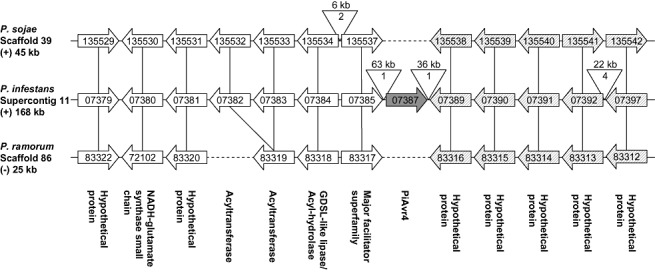
Schematic representation of the genomic region in Phytophthora infestans that carries PiAvr4 (gene model 07387) and regions with conserved synteny in P. sojae and P. ramorum. The sizes of the aligned regions are indicated, as well as the orientation of the regions in the respective scaffolds or contig (+ or −). The orthologous gene models are aligned and the predicted function is indicated. Gene models with a high similarity to transposon sequences are depicted in the inverted triangles, in which the number of transposon gene models is indicated. Of the gene clusters located downstream of PiAvr4, only five of the more than 30 gene models are shown (shaded arrows). The spacing between the gene models is not to scale. Broken lines are used to connect gene models, but do not represent the actual relative distance between gene models.
Table 1.
Gene models surrounding the PiAvr4 gene in Phytophthora infestans.
| Model | Start | Stop | +/−* | Predicted protein | Orthologue in P. ramorum | Orthologue in P. sojae |
|---|---|---|---|---|---|---|
| PITG_07379.1 | 259036 | 261015 | − | Unknown | Pr83322 | Ps135529 |
| PITG_07380.1 | 261154 | 262848 | − | NADH‐glutamate synthase small chain | Pr72102 | Ps135530 |
| PITG_07381.1 | 265234 | 267078 | − | Unknown | Pr83320 | Ps135531 |
| PITG_07382.1 | 290966 | 292351 | − | Acyltransferase | Pr83319 | Ps135532 |
| PITG_07383.1 | 293408 | 293362 | − | Acyltransferase | Pr83319 | Ps135533 |
| PITG_07384.1 | 295171 | 295112 | − | GDSL‐like lipase/acyl‐hydrolase | Pr83318 | Ps135534 |
| PITG_07385.1 | 295644 | 297048 | + | Major facilitator superfamily | Pr83317 | Ps135537 |
| PITG_07386.1 | 298302 | 298113 | − | Transposon | −† | − |
| PITG_07387.1 | 359782 | 360645 | + | PiAvr4 | − | Ps109418 |
| PITG_07388.1 | 367654 | 368802 | − | Transposon | − | N/A |
| PITG_07389.1 | 396383 | 397687 | − | Unknown | Pr83315 | Ps135538 |
| PITG_07390.1 | 398114 | 399417 | − | Unknown | Pr83314 | Ps135539 |
| PITG_07391.1 | 400069 | 401375 | − | Unknown | Pr83312 | Ps135540 |
| PITG_07392.1 | 402975 | 402920 | − | Unknown | Pr83315 | Ps135541 |
| PITG_07393.1 | 404429 | 403757 | − | Transposon | − | − |
| PITG_07394.1 | 406403 | 408042 | + | Transposon | − | − |
| PITG_07395.1 | 408697 | 408282 | − | Transposon | − | − |
| PITG_07396.1 | 421229 | 422366 | + | Transposon | − | − |
| PITG_07397.1 | 425226 | 426428 | + | Unknown | Pr83314 | Ps135542 |
Orientation of the predicted open reading frame.
No orthologue present.
Downstream of PiAvr4 is a large gene cluster that is conserved between the three species. The more than 30 genes in this cluster are highly similar (with a paralogue sequence identity of 85%), but have no homology to known genes. In this gene cluster, P. infestans has an indel block of 22 kb that carries four transposon gene models, but no Avh genes. In P. infestans, the closest PiAvr4 homologues are located on other scaffolds. PiAvh_38 and PiAvh_190, sharing over 90% sequence similarity but with less than 50% similarity to PiAvr4, are located on scaffolds 9 and 19, respectively, and Pi_Avh131, with similarity restricted to the N‐terminus, is located on scaffold 1 (Table S1, see Supporting Information). PiAvh_38 and PiAvh_190 are probably recently duplicated paralogues, one of which is located on a segmental duplication of 3 kb that settled elsewhere in the genome. Often such closely related paralogues are found on the same scaffold, as is the case for PiAvh_331 and PiAvh_313 which share 91% protein sequence similarity and are 75 kb apart, and the nearly identical genes PsAvh_192 and PsAvh_193 which are only 32 kb apart. Remarkably, in addition, the less divergent PsAvh_110 and PsAvh_191, the two closest PiAvr4 homologues in P. sojae and sharing 60% protein sequence similarity, are only 25 kb apart. In contrast with the clustering of anciently duplicated paralogues. such as elicitin genes (Jiang et al., 2006b) and sPLD‐like genes (Meijer and Govers, 2006), many of the more divergent Avh genes are often scattered over the genome (Jiang et al., 2008).
Conserved motifs in the C‐terminus of PiAvr4
To find conserved W, Y and L motifs in the C‐terminal domain of PiAvr4, we used the HMMs developed by Jiang et al. (2008). Three W motifs, named W1, W2 and W3, and a single Y motif were identified, but no L motifs (Fig. 3a; Table 2). The HMM scores of the W motifs in PiAvr4 vary. W2 and W3 have scores of 19.8 and 10.9, respectively, whereas the score of W1 is 0.6. For this HMM, a score of >10 is considered as strong (Jiang et al., 2008). In addition, the P. mirabilis Avr4 homologue PmirAvh4 carries three W motifs and a single Y motif with a low HMM score for W1. The HMM score of the PmirAvh4 W2 motif is highest, with a value of 20.3, whereas W3 has a value of only 6.7. PsAvh_110, the closest PiAvr4 homologue in P. sojae, has two W motifs and a single Y motif (Jiang et al., 2008). Compared with PiAvr4, the W1 motif appears to be absent in PsAvh_110. Motifs W2, W3 and Y, and the inter‐motif regions, are conserved between PiAvr4 and PsAvh_110, although the similarity is low (27% identity). Of the other Avr4 family members, three have no recognizable motifs in the C‐terminal domain, including that which is selected on the basis of similarity in the N‐terminus (PiAvh_131); eight contain one W motif and no Y, and four contain two W motifs with or without Y. Apart from PiAvr4, only one family member contains three W motifs (Table S1, see Supporting Information). The variable number of motifs among the homologues suggests that dynamic deletion or duplication of motifs plays a role in the diversification of the C‐terminal domains of RXLR‐dEER effectors.
Figure 3.
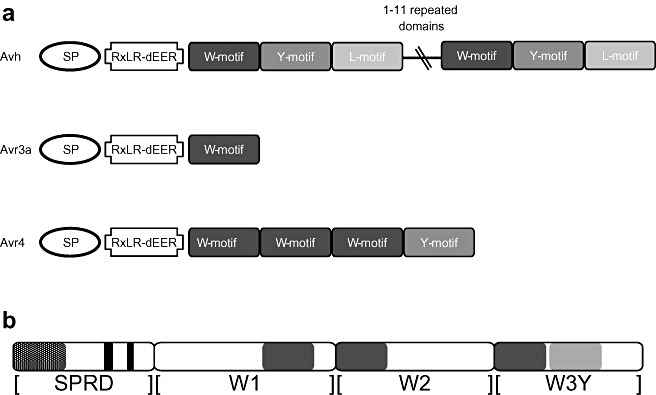
Oomycete Avh proteins have a modular structure. (a) Avh shows the basic components of an RXLR‐dEER protein, with an N‐terminal signal peptide (SP), the RXLR‐dEER domain and the C‐terminus with a variable number of modules that consist of W, Y and L motifs. PiAvr4 carries three W motifs and one Y motif. Avr3a, shown as an example of another Phytophthora infestans Avr protein, carries only a single W motif. (b) A schematic representation of the relative positions and sizes of the conserved motifs in PiAvr4 and the building blocks that were used to generate deletion constructs. Building block [SPRD] covers the SP (hatched) and the RXLR and dEER motifs (black). [W1], [W2] and [W3Y] cover the W motifs (dark grey) and the Y motif (light grey).
Table 2.
Hidden Markov Model (HMM) scores of the various domains and motifs in PiAvr4 and its closest homologues.

To further investigate the potential role of the W and Y motifs, we analysed the secondary structure of PiAvr4. In total, 12 α‐helices, ranging in size from five to 41 amino acids, are predicted in the C‐terminal region of PiAvr4 (http://www.sbg.bio.ic.ac.uk/phyre). Two of these are amphipathic α‐helices with hydrophobic and hydrophilic residues clustered on opposite sites; helix 5 ranges from residue 129 to 154 and covers parts of motifs W1 and W2, whereas helix 6 spans part of W2 and ranges from residue 159 to 199 (Fig. S2, see Supporting Information). In P. sojae Avr1b, three such amphipathic helices are found, and Dou et al. (2008a) showed that polymorphic residues are exclusively located in the hydrophilic sites of two helices coinciding with the W motif in Avr1b. Polymorphisms between PiAvr4 and PmirAvh4 do not specifically localize to either hydrophobic or hydrophilic sides of the helices and the secondary structure of both proteins is well conserved. However, part of the amphipathic structure in helix 5 is disrupted by the polymorphisms, resulting in a helix in PmirAvh4 that is 12 residues shorter. In addition, the predicted structures in helices 6, 8 and 9 are slightly different in PmirAvh4 compared with PiAvr4, but the amphipathic structures are not disrupted.
Elicitor activity is confined to restricted regions in the C‐terminus of PiAvr4
In a previous study (van Poppel et al., 2008), we expressed PiAvr4 in R4 potato plants and determined that the RXLR‐dEER domain is not required for elicitor activity of PiAvr4. To determine whether changes in any of the 27 amino acids in the C‐terminus of PiAvr4 that differ from PmirAvh4 abolish elicitor activity on R4 plants, we performed an agroinfection assay and transiently expressed PmirAvh4 in planta. R4 potato plants showed an HR at the inoculation site but, on Bintje, the potato cultivar lacking any known R gene, no response was observed (Fig. 4). Each of the three W domains of PmirAvh4 differ in terms of four amino acids compared with the corresponding W domain in PiAvr4 (Table 2), but apparently these amino acids can be changed without losing elicitor activity on R4 plants.
Figure 4.
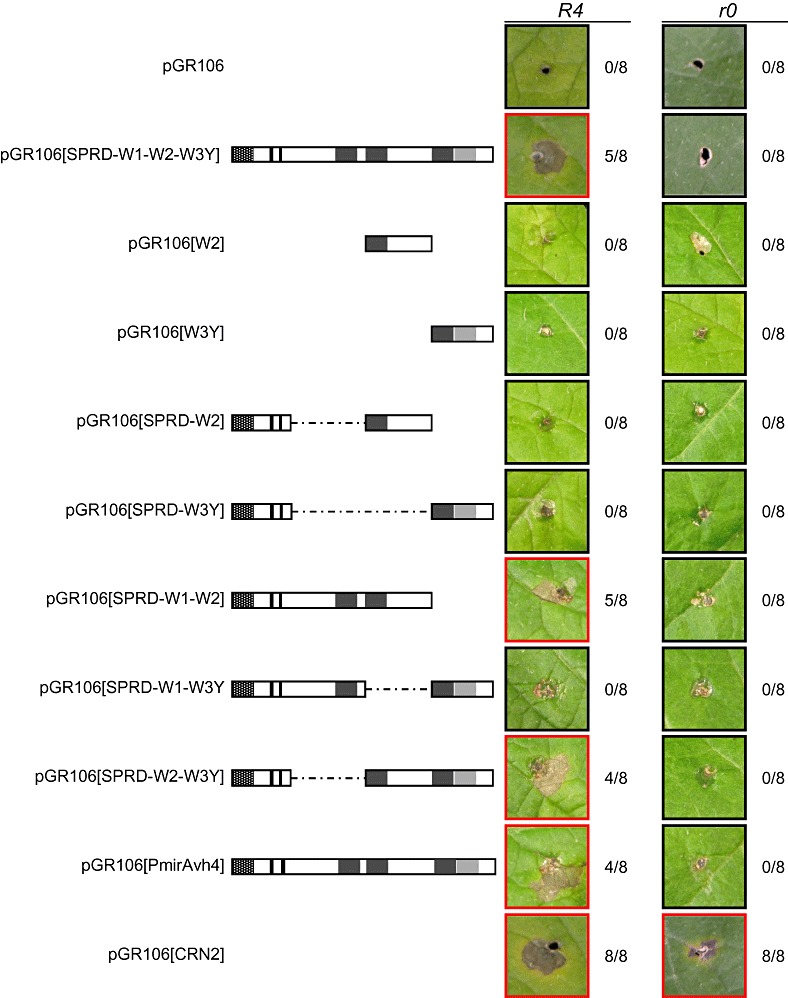
Agroinfection of potato line Cebeco44‐31‐5 (R4) and cultivar Bintje (r0) by toothpick inoculation with strains carrying pGR106 constructs as indicated. Broken lines connect the building blocks that are included in the constructs. The signal peptide is marked as a hatched box, the RXLR‐dEER motif in black and the W and Y motifs in dark and light grey, respectively. The numbers show the ratio of hypersensitive responses and the total number of toothpick inoculations in a typical experiment. Photographs were taken at 18 days post‐inoculation.
To investigate which part of the C‐terminus of PiAvr4 is responsible for elicitor activity, we generated various deletion constructs in a binary PVX vector for transient expression in planta. For the design of the constructs, the C‐terminal region of PiAvr4 was divided into three parts, each covering one of the W motifs. The constructs comprised the building blocks [SPRD], [W1], [W2] and [W3Y] (Fig. 3b). The control construct, encoding the full‐length PiAvr4, is thus pGR106[SPRD‐W1‐W2‐W3Y] (Fig. 3b and Table S2, see Supporting Information). In previous agroinfection assays, the full‐length pGR106‐Avr41–287 construct, but not the pGR106‐Avr425–287 construct, elicited a response on R4 potato plants (van Poppel et al., 2008), and therefore the SP sequence was included in all deletion constructs. In addition, we prepared two deletion constructs without SP (pGR106[W2] and pGR106[W3]). Despite several attempts, the cloning of both pGR106[W1] and pGR106[SPRD‐W1] was unsuccessful. As W1 has a low HMM score, no further attempts were made.
Toothpick inoculation with Agrobacterium tumefaciens carrying pGR106[SPRD‐W1‐W2] and pGR106[SPRD‐W2‐W3Y] resulted in an HR on R4 plants within 13 days post‐inoculation (dpi), whereas no response was visible on pGR106[SPRD‐W1‐W3Y] or any of the constructs carrying a single [W2] or [W3Y] building block (Fig. 4). None of the strains carrying the empty vector pGR106 showed a necrotic response, and neither did any of the inoculations on Bintje (r0). As positive controls, we used A. tumefaciens strains carrying pGR106‐CRN2; CRN2 is a P. infestans elicitor that induces general necrosis (Torto et al., 2003). If we assume that the proteins produced by the various deletion constructs are stable, even the shorter ones, such as [SPRD‐W2] and [SPRD‐W3Y], the results lead to the conclusion that the region carrying the W2 domain is required, but not sufficient, for the elicitation of HR on plants carrying R4.
DISCUSSION
A typical oomycete Avr protein consists of an SP, an RXLR‐dEER domain and a C‐terminal region. The SP is removed when the Avr protein is secreted by the pathogen and, as shown for P. infestans Avr3a (Whisson et al., 2007) and P. sojae Avr1b (Dou et al., 2008a), the RXLR‐dEER domain plays a crucial role in host cell targeting. The C‐terminus is the region that determines the avirulence or virulence function of the effector protein; it is responsible for recognition by host R proteins but, in the absence of the cognate R protein, it can often suppress cell death (Bos et al., 2006; Dou et al., 2008a). It is therefore not surprising that the C‐terminal regions of RXLR‐dEER effectors are under high positive selection (Jiang et al., 2008; Win et al., 2007). Nevertheless, more than one‐half of these C‐terminal regions contain conserved motifs, named W, Y and L (Jiang et al., 2008). The fact that these motifs are retained suggests that they are important for the function of RXLR‐dEER effectors. When assuming that R proteins recognize conserved regions in Avr proteins, the W, Y and L motifs probably encounter a strong host‐driven positive selection. Indeed, most variation in the P. sojae effector Avr1b was found in the W and L motifs and in a so‐called K motif. Moreover, mutations in conserved residues in these motifs abolished elicitor activity, avirulence function and suppression of cell death (Dou et al., 2008a).
In this article, we have shown that the P. infestans RXLR‐dEER effector PiAvr4 contains a C‐terminal region with three W motifs and one Y motif. In addition, most of the PiAvr4 homologues contain W motifs, but usually less than three. The closest homologue is PmirAvh4, obtained from the sibling species P. mirabilis. PmirAvh4 has the same number of motifs and 89% similarity in the C‐terminal region. Sequences identical to PmirAvh4 were found in two other sibling species, P. phaseoli and P. ipomoeae, but not in P. infestans (R.H.Y. Jiang, unpublished data), thus suggesting that PmirAvh4 is the ancestor of PiAvr4. Phytophthora mirabilis is a pathogen of Mirabilis jalapa, the four o'clock flower, and not pathogenic on potato (Grünwald and Flier, 2005). To address the question of whether PmirAvh4 is recognized by potato R4, we determined its effector activity by transient expression in R4 plants. In a previous study, we have shown that agroinfection with A. tumefaciens strains carrying a full‐length PiAvr4 construct in a binary PVX vector results in an HR on R4 plants (van Poppel et al., 2008), whereas constructs of PiAvr4 lacking the SP sequence do not elicit a response on R4 plants. In agroinfiltration assays, the difference between constructs plus and minus the SP sequence was less evident, but in all cases a full‐length PiAvr4 construct triggered necrotic responses specifically on R4 plants and not on cv. Bintje (r0). As deletion of the RXLR‐dEER domain did not change the response, we concluded that the elicitor activity of PiAvr4 is restricted to the C‐terminal region (van Poppel et al., 2008). In this study, we cloned the full‐length PmirAvh4 gene in the binary PVX vector and found that, similar to PiAvr4, PmirAvh4 is recognized by R4. This shows that potato R4 recognizes an ancestral RXLR‐dEER effector and that the 27 amino acids that have changed since the divergence of P. infestans from its sibling species are not essential determinants for recognition. Apparently, more substantial or more specific mutations are required to evade recognition by potato R4. The finding that all P. infestans isolates that are virulent on potato R4 carry a frameshift mutation in PiAvr4 (van Poppel et al., 2008) suggests that P. infestans uses a rather robust mechanism to remove PiAvr4 activity. More subtle mechanisms are specific point mutations, as found for P. infestans Avr3a and the Cladosporium fulvum Avr4 gene. For the latter, recognition by the tomato R protein Cf‐4 is abolished by a single amino acid change in Avr4 (Joosten et al., 1994). In the case of Avr3a, isolates that are virulent on R3a plants have an intact open reading frame (ORF) at the Avr3a locus. This ORF encodes an effector that has two specific point mutations, but lacks elicitor activity on R3a plants (Armstrong et al., 2005). Similarly, ATR13 in H. parasitica has four amino acid residues that determine RPP13‐mediated resistance (Allen et al., 2008). Mutations in these residues abolish elicitor activity.
The frameshift mutations in PiAvr4 in virulent isolates always occur at two fixed positions, and the proteins encoded by the remaining ORF are either 17 or 92 amino acids in size (van Poppel et al., 2008). The latter covers the RXLR‐dEER domain and a small part of the C‐terminal domain, but this is not sufficient to trigger HR. To determine which part of the C‐terminal domain has elicitor activity, we tested various deletion constructs. The data show that the region containing the W2 motif is essential, but not sufficient; flanking regions, either upstream or downstream, are necessary to elicit an HR. As the W1 and W3 motifs have lower HMM scores than W2, they may be less important for the recognition itself. Possibly, the flanking regions provide stability to the central region that comprises W2. Unfortunately, we did not incorporate tags into the deletion constructs and therefore were unable to analyse protein production and/or stability.
In their study on P. sojae Avr1b, Dou et al. (2008a) demonstrated that mutations in the W domain cause a loss of avirulence function. Some of the field isolates virulent on Rps1b soybean plants carry alleles with mutations at these positions in the W domain, which explains their virulent phenotype. Similarly, with the two naturally occurring alleles of P. infestans Avr3a, the identity of two key amino acids as determinants of avirulence was uncovered (Armstrong et al., 2005). In contrast, analysis of P. infestans field isolates virulent on R4 plants has not revealed which amino acids are the potential determinants of avirulence function. We have found several point mutations in virulent alleles but, as there is always a frameshift mutation in the 5′ region of the gene at a fixed position, we assume that these point mutations were introduced after the frameshift mutation and at random without any selection pressure (van Poppel et al., 2008).
In the genomes of Phytophthora spp., Avh genes are mostly located in regions that show genome rearrangements and mark breakpoints of conserved synteny (Jiang et al., 2008). PiAvr4 follows this pattern; the genomic region comprising PiAvr4 has conserved synteny with genomic regions in both P. sojae and P. ramorum, but the conserved synteny is disrupted by PiAvr4. The 45‐kb region in P. sojae and the 25‐kb region in P. ramorum, which match 168 kb in P. infestans, lack Avh genes. The size differences are a result of transposon‐like sequences that are more abundant in P. infestans and dispersed throughout this region, and a 100‐kb indel that carries PiAvr4 flanked by transposon‐derived sequences. In addition, P. sojae Avr1b‐1 is located on a 50‐kb indel that is absent in the syntenic region in P. ramorum (Jiang et al., 2006a). Overall, Avh genes in Phytophthora are often associated with retroelements, such as transposons. As observed with several of the PiAvr4 homologues, new paralogues are often clustered, whereas other older paralogue members are scattered around the genome. This scattering may prevent homogenization via illegitimate recombination between duplicated genes and may contribute to the rapid divergence of the Avh gene family (Jiang et al., 2008).
EXPERIMENTAL PROCEDURES
Genomic sequences
The genomic sequences and gene models of P. sojae (version 1.1) and P. ramorum (version 1.1) were retrieved from the website of the DOE Joint Genome Institute (http://genome.jgi‐psf.org/) and of P. infestans (version 1.0) from the website of the Broad Institute (http://www.broad.mit.edu/). The genomic sequences of P. capsici and H. parasitica were accessed at http://genomeportal.jgi‐psf.org/PhycaF7 and http://vmd.vbi.vt.edu/, respectively.
Identification of C‐terminal motifs by HMMs
Using the program HMMER 2.3.2 (15) (http://hmmer.janelia.org/), three HMMs were built from the RXLR‐dEER effectors that carry conserved C‐terminal motifs (Jiang et al., 2008). The sequences of the W, Y and L motifs that were used to build the HMMs are available in Table S3 (see Supporting Information). HMM scores of >10 were considered as strong. One HMM was built from the RXLR‐dEER motifs, with the variable spacing arbitrarily placed in between. The RXLR‐dEER motif is defined as the occurrence of the string RXLR together with the trailing acidic motif (containing more than 10% D or E residues). The HMM building method is very similar to that described by Jiang et al. (2008). To increase the sensitivity of a database search, the model was calibrated by ‘hmmcalibrate’ to give an empirical E value calculation according to the HMM as suggested by the program instructions. Motif searches were performed with these four HMMs on the total set of RXLR protein sequences from P. infestans, P. sojae and P. ramorum.
Homologue search
The entire sets of RXLR‐dEER effectors of P. infestans (562), P. sojae (385) and P. ramorum (370) were gathered to make an effector pool. PiAvr4 was used to perform blastp against the effector pool and to identify direct homologues. Similar blastp searches were performed with these direct homologues to discover more RXLR‐dEER family members, and a few additional, most similar homologues were included in the Avr4 family.
Orthologue search
The location of PiAvr4 was determined by a blastn search against the complete P. infestans genomic sequence. Gene models surrounding PiAvr4 were selected and used for a blast search of the complete genomic sequences of P. sojae and P. ramorum. Gene models with the best reciprocal blast hit were assigned as orthologues. Because transposon derived sequences rarely have homologues in syntenic regions, and because of the repetitive nature of transposon sequences, these gene models were excluded from the analysis.
Binary PVX constructs and agroinfection
To express PiAvr4 in R4 and r0 potato plants, Cebeco44‐31‐5 and cv. Bintje, respectively, binary PVX constructs were prepared in the vector pGR106 (Jones et al., 1999). BAC subclone pSKA23 (van Poppel et al., 2008) was used as a template to amplify specific parts of the PiAvr4 gene. Primers ClaIAvr4F and NotIAvr4R were used to amplify a PiAvr4 homologue from P. mirabilis isolate PIC99111 (Flier et al., 2002), and designated PmirAvh4. The primers used contained appropriate restriction sites or sequences for overlap PCR (Table S4, see Supporting Information). For cloning purposes, a ClaI restriction site was incorporated into the forward primer of the construct and a NotI site into the reverse primer. The amplicons obtained were digested by the appropriate enzymes and cloned into pGR106. The binary PVX constructs were then transformed into A. tumefaciens strain GV3101 for agroinfection and strain AGL1 for agroinfiltration assays. The integrity of the constructs was checked by sequencing. The DNA sequence of PmirAvh4 was deposited in GenBank (accession number FJ951909).
Agroinfection assays on potato were performed as described previously (Vleeshouwers et al., 2006). Briefly, A. tumefaciens strains containing the binary PVX constructs were grown for 2 days on LBman agar medium. Toothpicks were used to transfer bacteria to the leaves and to pierce the leaf, creating wound tissue. Responses were monitored for up to 4 weeks post‐inoculation.
Supporting information
Fig. S1 Alignment of PiAvr4 and PmirAvr4.
Fig. S2 Predicted α‐helices in PiAvr4 and helical wheel projections of the amphipathic helices.
Table S1 Sizes of PiAvr4 family members and occurrence of motifs in their C‐terminal domains.
Table S2 Constructs used in this study.
Table S3 W, Y and L motifs used to build a Hidden Markov Model (HMM) with HMMER 2.3.2.
Table S4 Polymerase chain reaction (PCR) primers used for cloning.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
We thank Henk Smid and Bert Essenstam for support in the glasshouse, Harold Meijer for valuable discussions and Pierre de Wit for critical reading of the manuscript. We also thank Wilbert Flier for providing P. mirabilis strain PIC99111. This project was (co)financed by the Centre for BioSystems Genomics (CBSG), which is part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research. JS was financially supported by a fellowship from the European Union project BioExploit (FOOD‐CT‐2005‐513959).
REFERENCES
- Allen, R.L. , Bittner‐Eddy, P.D. , Grenville‐Briggs, L.J. , Meitz, J.C. , Rehmany, A.P. , Rose, L.E. and Beynon, J.L. (2004) Host–parasite coevolutionary conflict between arabidopsis and downy mildew. Science, 306, 1957–1960. [DOI] [PubMed] [Google Scholar]
- Allen, R.L. , Meitz, J.C. , Baumber, R.E. , Hall, S.A. , Lee, S.C. , Rose, L.E. and Beynon, J.L. (2008) Natural variation reveals key amino acids in a downy mildew effector that alters recognition specificity by an arabidopsis resistance gene. Mol. Plant Pathol. 9, 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong, M.R. , Whisson, S.C. , Pritchard, L. , Bos, J.I.B. , Venter, E. , Avrova, A.O. , Rehmany, A.P. , Bohme, U. , Brooks, K. , Cherevach, I. , Hamlin, N. , White, B. , Fraser, A. , Lord, A. , Quail, M.A. , Churcher, C. , Hall, N. , Berriman, M. , Huang, S. , Kamoun, S. , Beynon, J.L. and Birch, P.R.J. (2005) An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc. Natl. Acad. Sci. USA, 102, 7766–7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, W. , Mastenbroek, C. , Mills, W.R. and Peterson, L.C. (1953) A proposal for an international nomenclature of races of Phytophthora infestans and of genes controlling immunity in Solanum demissum derivates. Euphytica, 2, 173–179. [Google Scholar]
- Bos, J.I. , Kanneganti, T.D. , Young, C. , Cakir, C. , Huitema, E. , Win, J. , Armstrong, M. , Birch, P.R.J. and Kamoun, S. (2006) The C‐terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a‐mediated hypersensitivity and suppress INF1‐induced cell death in Nicotiana benthamiana . Plant J. 48, 165–176. [DOI] [PubMed] [Google Scholar]
- Bouwmeester, K. , Van Poppel, P.M.J.A. and Govers, F. (2009) Genome biology cracks enigmas of oomycete plant pathogens In: Molecular Aspects of Plant Disease Resistance, Vol. 34 (Parker J.E., ed.), pp. 102–134. Oxford: Wiley‐Blackwell. [Google Scholar]
- Dou, D. , Kale, S.D. , Wang, X. , Chen, Y. , Wang, Q. , Wang, X. , Jiang, R.H.Y. , Arredondo, F.D. , Anderson, R.G. , Thakur, P.B. , McDowell, J.M. , Wang, Y. and Tyler, B.M. (2008a) Conserved C‐terminal motifs required for avirulence and suppression of cell death by Phytophthora sojae effector Avr1b. Plant Cell, 20, 1118–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou, D. , Kale, S.D. , Wang, X. , Jiang, R.H.Y. , Bruce, N.A. , Arredondo, F.D. , Zhang, X. and Tyler, B.M. (2008b) RXLR‐mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen‐encoded machinery. Plant Cell, 20, 1930–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flier, W.G. , Grünwald, N.J. , Kroon, L.P.N.M. , Van Den Bosch, T.B.M. , Garay‐Serrano, E. , Lozoya‐Saldaña, H. , Bonants, P.J.M. and Turkensteen, L.J. (2002) Phytophthora ipomoeae sp. nov., a new homothallic species causing leaf blight on Ipomoea longipedunculata in the Toluca Valley of central Mexico. Mycol. Res. 106, 848–856. [Google Scholar]
- Govers, F. and Gijzen, M. (2006) Phytophthora genomics: the plant destroyers' genome decoded. Mol. Plant–Microbe Interact. 19, 1295–1301. [DOI] [PubMed] [Google Scholar]
- Grünwald, N.J. and Flier, W.G. (2005) The biology of Phytophthora infestans at its center of origin. Annu. Rev. Phytopathol. 43, 171–190. [DOI] [PubMed] [Google Scholar]
- Jiang, R.H.Y. , Tripathy, S. , Govers, F. and Tyler, B.M. (2008) RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc. Natl. Acad. Sci. USA, 105, 4874–4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, R.H.Y. , Tyler, B.M. and Govers, F. (2006a) Comparative analysis of Phytophthora genes encoding secreted proteins reveals conserved synteny and lineage‐specific gene duplications and deletions. Mol. Plant–Microbe. Interact. 19, 1311–1321. [DOI] [PubMed] [Google Scholar]
- Jiang, R.H.Y. , Tyler, B.M. , Whisson, S.C. , Hardham, A.R. and Govers, F. (2006b) Ancient origin of elicitin gene clusters in Phytophthora genomes. Molec. Biol. Evol. 23, 338–351. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Jones, L. , Hamilton, A.J. , Voinnet, O. , Thomas, C.L. , Maule, A.J. and Baulcombe, D.C. (1999) RNA–DNA interactions and DNA methylation in post‐transcriptional gene silencing. Plant Cell, 11, 2291–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten, M.H.A.J. , Cozijnsen, T.J. and De Wit, P.J.G.M. (1994) Host‐resistance to a fungal tomato pathogen lost by a single base‐pair change in an avirulence gene. Nature, 367, 384–386. [DOI] [PubMed] [Google Scholar]
- Meijer, H.J.G. and Govers, F. (2006) Genomewide analysis of phospholipid signaling genes in Phytophthora spp.: novelties and a missing link. Mol. Plant–Microbe. Interact. 19, 1337–1347. [DOI] [PubMed] [Google Scholar]
- Van Poppel, P.M.J.A. , Guo, J. , Van de Vondervoort, P.J.I. , Jung, M.W.M. , Birch, P.R.J. , Whisson, S.C. and Govers, F. (2008) The Phytophthora infestans avirulence gene Avr4 encodes an RXLR‐dEER effector. Mol. Plant–Microbe. Interact. 21, 1460–1470. [DOI] [PubMed] [Google Scholar]
- Qutob, D. , Kamoun, S. and Gijzen, M. (2002) Expression of a Phytophthora sojae necrosis‐inducing protein occurs during transition from biotrophy to necrotrophy. Plant J. 32, 361–373. [DOI] [PubMed] [Google Scholar]
- Rehmany, A.P. , Gordon, A. , Rose, L.E. , Allen, R.L. , Armstrong, M.R. , Whisson, S.C. , Kamoun, S. , Tyler, B.M. , Birch, P.R.J. and Beynon, J.L. (2005) Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell, 17, 1839–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torto, T.A. , Li, S. , Styer, A. , Huitema, E. , Testa, A. , Gow, N.A.R. , Van West, P. and Kamoun, S. (2003) EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora . Genome Res. 13, 1675–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleeshouwers, V.G.A.A. , Driesprong, J.‐D. , Kamphuis, L.G. , Torto‐Alalibo, T.A. , Van’t Slot, K.A.E. , Govers, F. , Visser, R.G.F. , Jacobsen, E. and Kamoun, S. (2006) Agroinfection‐based high‐throughput screening reveals specific recognition of INF elicitins in Solanum . Mol. Plant Pathol. 7, 499–510. [DOI] [PubMed] [Google Scholar]
- Whisson, S.C. , Boevink, P.C. , Moleleki, L. , Avrova, A.O. , Morales, J.G. , Gilroy, E.M. , Armstrong, M.R. , Grouffaud, S. , Van West, P. , Chapman, S. , Hein, I. , Toth, I.K. , Pritchard, L. and Birch, P.R.J. (2007) A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature, 450, 115–118. [DOI] [PubMed] [Google Scholar]
- Win, J. , Morgan, W. , Bos, J. , Krasileva, K.V. , Cano, L.M. , Chaparro‐Garcia, A. , Ammar, R. , Staskawicz, B.J. and Kamoun, S. (2007) Adaptive evolution has targeted the C‐terminal domain of the RXLR effectors of plant pathogenic oomycetes. Plant Cell, 19, 2349–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Alignment of PiAvr4 and PmirAvr4.
Fig. S2 Predicted α‐helices in PiAvr4 and helical wheel projections of the amphipathic helices.
Table S1 Sizes of PiAvr4 family members and occurrence of motifs in their C‐terminal domains.
Table S2 Constructs used in this study.
Table S3 W, Y and L motifs used to build a Hidden Markov Model (HMM) with HMMER 2.3.2.
Table S4 Polymerase chain reaction (PCR) primers used for cloning.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item


