Figure 1.
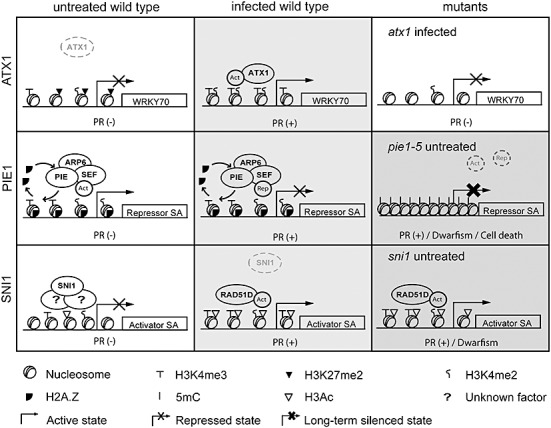
Model for Arabidopsis Trithorax 1 (ATX1)‐, Photoperiod‐Independent Early flowering 1 (PIE1)‐ and Suppressor of NPR1, Inducible 1 (SNI1)‐mediated epigenetic control of the salicylic acid (SA) pathway. Top: After infection, ATX1 binds the WRKY70 nucleosomes and mediates the trimethylation of H3K4. This modification recruits transcriptional activators and leads to WRKY70 expression and subsequent pathogenesis‐related (PR) gene induction. The absence of H3K4‐me3 on the WRKY70 nucleosomes in the atx1 mutant impairs the activation of WRKY70 after infection. Middle: The Swr1 complex (PIE/SEF/ARP6) introduces H2A.Z at the promoter of a gene encoding a repressor of the SA pathway (Repressor SA). H2A.Z and methylated H3K4 maintain the accessibility of this promoter to transcriptional regulators. Activators (Act) or repressors (Rep) bind this region under basal or infection conditions, respectively. Although the removal of H2A.Z may take place during transcription (curved arrows), this modification protects the promoter from cytosine methylation (5mC). The absence of H2A.Z in pie1‐5 plants leads to chromatin compaction, DNA methylation and gene silencing, and the subsequent activation of defence mechanisms in noninfected tissues that causes growth retardation. Bottom: In basal conditions, SNI1 prevents the expression of positive regulators of the SA pathway (Activator SA) by maintaining local histone modifications that reduce chromatin accessibility. After infection, SNI1 is released from the promoter and chromatin is relaxed by RAD51D activity. SNI1 may affect chromatin structure through interactions with other proteins of unknown nature. The grey scale illustrates the degrees in SA pathway activation (white, no activation). Dwarfism and cell death phenotypes most probably result from constitutive activation of the pathway.
