Figure 2.
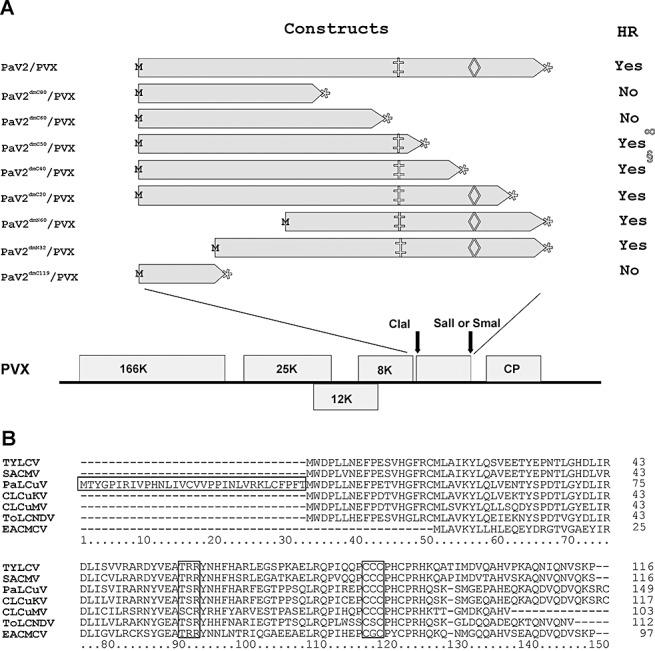
Constructs used in the analysis and comparison of (A)V2 sequences. (A) Potato virus X (PVX)‐based expression cassettes of V2 of PaLCuV. The deletion mutants of V2 of PaLCuV are indicated with their amino acid coordinates. M and  are the start and stop codons, respectively. The positions of the putative protein kinase C motif (‡) and CxC motif (◊) are indicated. HR, hypersensitive response. (B) Alignment of the predicted amino acid sequences of the V2 proteins of PaLCuV‐PK[PK:Cot:02] (AJ436992) and CLCuKoV‐Fai[PK:Fai1] (AJ496286) with the V2 proteins of other selected begomoviruses: Tomato yellow leaf curl virus (TYLCV) (AF260331), South African cassava mosaic virus (SACMV‐[ZW:Muz]; AJ575560), Cotton leaf curl Multan virus (CLCuMV‐Fai[PK:Fai2]; AJ496287), Tomato leaf curl New Delhi virus (ToLCNDV‐IN[IN:ND:Svr:92]; U15015) and East African cassava mosaic Cameroon virus (EACMCV‐CM[CM:98]; AF112354). Gaps (‐) were introduced to optimize the alignment. The position of a predicted putative protein kinase C motif (TxR; coordinates 91–93 of the alignment) and the CxC motif (coordinates 116–118 of the alignment) mutated by Padidam et al., (1996) and Zrachya et al., (2007) are shown. The precise position of the initiation methionine of (A)V2 has not been determined. The V2 open reading frame of PaLCV has the capacity to encode an additional 32 amino acids at the N‐terminus in comparison with the other viruses.
are the start and stop codons, respectively. The positions of the putative protein kinase C motif (‡) and CxC motif (◊) are indicated. HR, hypersensitive response. (B) Alignment of the predicted amino acid sequences of the V2 proteins of PaLCuV‐PK[PK:Cot:02] (AJ436992) and CLCuKoV‐Fai[PK:Fai1] (AJ496286) with the V2 proteins of other selected begomoviruses: Tomato yellow leaf curl virus (TYLCV) (AF260331), South African cassava mosaic virus (SACMV‐[ZW:Muz]; AJ575560), Cotton leaf curl Multan virus (CLCuMV‐Fai[PK:Fai2]; AJ496287), Tomato leaf curl New Delhi virus (ToLCNDV‐IN[IN:ND:Svr:92]; U15015) and East African cassava mosaic Cameroon virus (EACMCV‐CM[CM:98]; AF112354). Gaps (‐) were introduced to optimize the alignment. The position of a predicted putative protein kinase C motif (TxR; coordinates 91–93 of the alignment) and the CxC motif (coordinates 116–118 of the alignment) mutated by Padidam et al., (1996) and Zrachya et al., (2007) are shown. The precise position of the initiation methionine of (A)V2 has not been determined. The V2 open reading frame of PaLCV has the capacity to encode an additional 32 amino acids at the N‐terminus in comparison with the other viruses.
