Abstract
Chagas disease was described by Carlos Chagas, who first identified the parasite Trypanosoma cruzi from a 2-year-old girl called Berenice. Many T. cruzi sequencing projects based on short reads have demonstrated that genome assembly and downstream comparative analyses are extremely challenging in this species, given that half of its genome is composed of repetitive sequences. Here, we report de novo assemblies, annotation, and comparative analyses of the Berenice strain using a combination of Illumina short reads and MinION long reads. Our work demonstrates that Nanopore sequencing improves T. cruzi assembly contiguity and increases the assembly size in ∼16 Mb. Specifically, we found that assembly improvement also refines the completeness of coding regions for both single-copy genes and repetitive transposable elements. Beyond its historical and epidemiological importance, Berenice constitutes a fundamental resource because it now constitutes a high-quality assembly available for TcII (clade C), a prevalent lineage causing human infections in South America. The availability of Berenice genome expands the known genetic diversity of these parasites and reinforces the idea that T. cruzi is intraspecifically divided in three main clades. Finally, this work represents the introduction of Nanopore technology to resolve complex protozoan genomes, supporting its subsequent application for improving trypanosomatid and other highly repetitive genomes.
Keywords: Trypanosoma cruzi, Berenice, hybrid assembly, protozoan parasites, Chagas disease, Oxford Nanopore Technologies
Introduction
The Oxford Nanopore sequencing technology is useful for assembling genomes that are rich in repetitive elements because its long reads can span entire tandems of repeats and anchor them to uniquely occurring segments of the genome, resolving these complex regions and improving contiguity. However, the still high error rates of this technology demands considerable amounts of data and intensive computation to build entire genomes just using long reads. Conversely, hybrid strategies that combine error-prone long reads with much more accurate Illumina short reads represent a more convenient approach for enhancing genome completeness. Indeed, several organisms ranging from bacteria (Wick et al. 2017) to vertebrates (Tan et al. 2018) have been recently sequenced using a combination of Nanopore and Illumina reads. However, this strategy has not been implemented so far to resolve protozoan genomes.
Trypanosoma cruzi is a protozoan parasite belonging to the order Kinetoplastida that causes Chagas disease, also known as American Trypanosomiasis, a neglected parasitic disease that affects 6–7 million people worldwide and is transmitted to humans and animals mainly by Triatomine insect vectors (Deane 1964; WHO 2017). Chagas disease recently emerged in nonendemic regions such as Western Europe, Australia, Japan, Canada, and the United States due to widespread immigration, however its highest incidence is observed in Latin American countries where the parasite is endemic (Rassi et al. 2010). Indeed, it was first diagnosed in Brazil more than one century ago by Carlos Chagas when he examined the 2-year-old girl Berenice Soares (Chagas 1909), who developed the asymptomatic form of the disease (de Lana et al. 1996). The archetypal T. cruzi strain originally isolated from this case (Salgado et al. 1962) represents the oldest known record for this pathogenic parasite, and own invaluable historical, cultural, and epidemiological importance. The Berenice strain belongs to TcII and has been characterized in many aspects but has not been whole-genome sequenced by any technology so far.
Here, we report the whole-genome sequence, annotation, and comparative analysis of the Berenice strain isolated by Salgado et al. (1962) using a combination of Illumina short reads and Nanopore long reads, providing a useful genetic resource for the community working with parasite genomes. Importantly, we demonstrate that a single run using the MinION sequencer based on a straightforward 10-min library preparation protocol allows a 67-fold increase in genome contiguity and improves genome completeness by 28% when compared with short-read-only assemblies. Our results show that hybrid assembly strategies using MinION are effective when dealing with complex protozoan genomes like T. cruzi.
Materials and Methods
Library Preparation, Genome Sequencing, and Assembly
Genomic libraries were prepared with the Nextera XT Library Prep Kit (Illumina, 15032354) and Rapid Sequencing Kit (Nanopore, SQK-RAD004). Illumina and Nanopore libraries were sequenced in MiSeq and MinION platforms, producing 12,589,973 paired-end short reads and 265,221 long reads, respectively. Integrity of Illumina libraries were analyzed using 2100 Bioanalyzer (Agilent) and quantified using Qubit dsDNA HS Assay Kit. Berenice genome assembly was performed using Illumina reads (Illumina genome assembly) and mixing Illumina and Nanopore reads (Hybrid genome assembly) with MaSuRCA using default parameters (Zimin et al. 2013, 2017).
Comparison of Genome Assemblies
For genome assembly comparisons, Illumina and Nanopore reads were aligned to Berenice genome assembled with both reads using minimap2 v2.10-r784 (Li 2018) with default parameters. Per-base genome coverage was calculated using bedtools v2.26.0 (Quinlan and Hall 2010) and samplot (Belyeu et al. 2018) was used for rendering the sequencing coverage in specific genomic regions. Completeness of genome coding regions was assessed using BUSCO v3.0.2 (Simão et al. 2015) with the eukaryotic and protist databases.
Genome Annotation
In order to annotate the coding sequences, the annotated proteins of 41 protozoan parasites genomes were obtained from TriTrypDB release 38 (http://tritrypdb.org/). Otherwise, all open reading frames longer than 150 amino acids were retrieved between start and stop codon using getorf from the EMBOSS suite (Rice et al. 2000) in both the hybrid and Illumina assemblies. Homologous genes were recovered using BLAST+ BlastP (Camacho et al. 2009), with alignment coverage >80%, identity percentage >80%, and an e-value threshold of 1e-10. Rfam release 13 (Nawrocki et al. 2015) and Infernal v1.1.1 (Nawrocki and Eddy 2013) were used for the annotation of noncoding genes as it was previously described (Kalvari et al. 2018). For tRNAs, tRNAscan-SE v.1.3.1 (Lowe and Chan 2016) was used with the eukaryotic model. Transposable elements were annotated using BLAST+ BlastN (Camacho et al. 2009) and tandem repeats were annotated using Tandem Repeat Finder v4.09 (Benson 1999).
Phylogenetic Analysis
Complete nucleotide sequences of L1Tc transposable elements were used to perform phylogenetic analyses. Sequences retrieved from six genomes were aligned using MAFFT v7.310 (Katoh and Standley 2013) with the L-ins-i option. A maximum-likelihood phylogenetic tree was reconstructed using PhyML v20120412 (Guindon et al. 2010) using the best-fitted model GTR selected with ModelGenerator v0.85 (Benson 1999).
Results and Discussion
Trypanosoma cruzi is the causative agent of Chagas disease, an important neglected tropical disease that affects about 6–7 million people worldwide (WHO 2017). Here, we report the complete genome sequence of T. cruzi strain Berenice, isolated from the patient in which Dr Carlos Chagas described the disease (Chagas 1909). This represents the first trypanosomatid parasite genome generated using a hybrid assembly strategy by combining Illumina short reads and Nanopore long reads. Even though trypanosomatid genomes are small, their assembly and annotation have been challenging due to the abundance of repetitive sequences including the 195-bp satellite, tandem repeats, and multigene families (El-Sayed, Myler, Bartholomeu, et al. 2005; Berná et al. 2018; Pita et al. 2019). In fact, when “tritryp” genomes were sequenced in 2005, T. cruzi genome assembly remained highly fragmented (El-Sayed, Myler, Blandin, et al. 2005), hampering highly precise comparative genomics. However, the recent advent of long-read sequencing technologies is allowing us to overcome these limitations. Long-read sequencing using PacBio has been proven useful to improve the quality of T. cruzi genome assemblies (Berná et al. 2018; Callejas-Hernández et al. 2018); however, the innovative Nanopore technology has been not implemented to sequence trypanosomatid genomes so far, despite presenting several comparative advantages over PacBio. Nanopore is cheaper, easy to use in any laboratory, requires less amount of genomic DNA, and sequencing yield can be monitored in real-time. Additionally, Nanopore offers countless possibilities for library preparation including quick, straightforward protocols. Indeed, here we show that a 10-min library preparation protocol followed by 12 h of Nanopore 1D sequencing significantly improves assembly contiguity and annotation, demonstrating the usefulness of this technology to resolve highly complex parasite genomes.
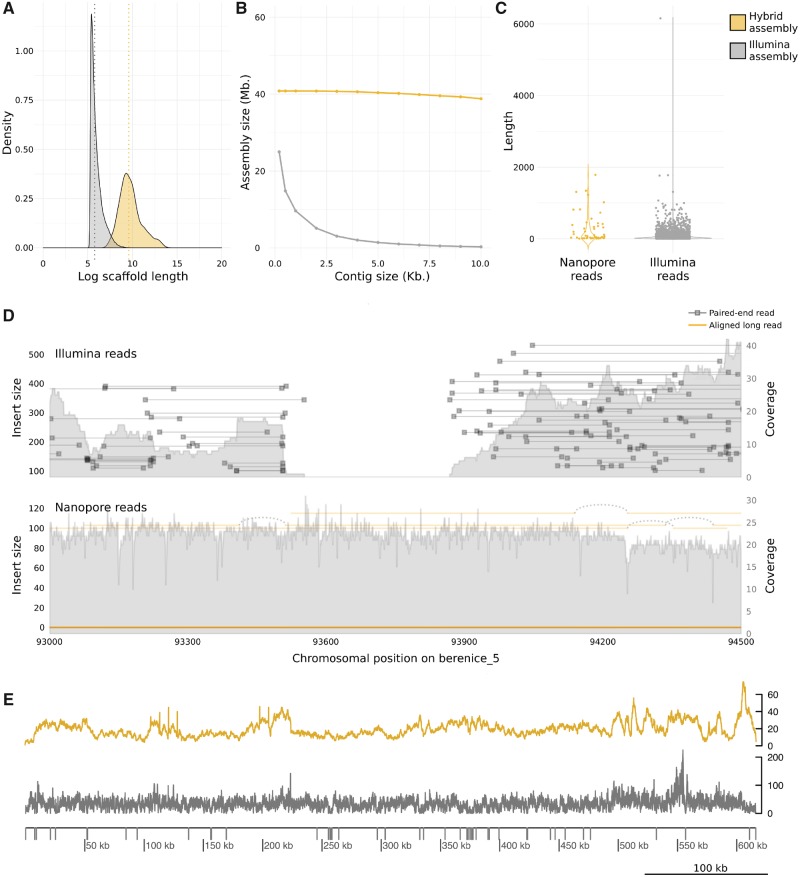
We whole-genome sequenced T. cruzi strain Berenice using Illumina 150-bp pair-end short reads and Nanopore 1D long reads (supplementary table 1, Supplementary Material online). Then, we produced two genome assemblies, one just using the short reads from Illumina (hereinafter referred as the Illumina assembly) and the other by combining Illumina short reads with Nanopore long reads (hereinafter referred as the hybrid assembly). Figure 1A shows a 46-fold improvement in median scaffold size in the hybrid assembly. This improvement is also evident by a 51-fold decrease in scaffold number (from ∼47,000 scaffolds with a maximum length of ∼26 kb in the Illumina assembly to ∼900 scaffolds with a maximum length of ∼1 Mb in the hybrid assembly), and a ∼16-Mb increase in assembly size product of improved resolution of repeated regions (supplementary table 1, Supplementary Material online). Also, the cumulative hybrid assembly size is kept practically unchanged around ∼40 Mb when considering scaffolds of increasing size, evidencing insignificant contribution of small scaffolds to the whole assembly. On the contrary, the cumulative size of the Illumina assembly rapidly tends to zero when considering longer scaffolds evidencing an extremely fragmented assembly (fig. 1B).
Fig. 1.
—Nanopore sequencing improves Trypanosoma cruzi assembly contiguity and size. (A) Scaffolds length distribution. Dotted lines indicates median of lengths. Median of scaffold length in hybrid assembly: 14,661. Median of scaffold length in Illumina assembly: 321. (B) Cumulative assemblies size. (C) Coverage zero regions (no read alignment in at least six consecutive positions) observed when Nanopore or Illumina reads were aligned to the hybrid assembly in order to assess the contribution of both technologies to the assembly contiguity. (D) Sequencing coverage and insert size from 93- to 94.5-kb positions of scaffold berenice_5 from hybrid assembly are plotted. (E) Per-base genome coverage of scaffold 4 of hybrid assembly. Coverage zero regions are plotted as gray bars over the exe and all were observed when Illumina reads were aligned to hybrid assembly.
To evaluate the contribution of Illumina and Nanopore data to close gaps, we separately aligned both types of reads to the hybrid assembly. The longest region where the coverage is zero (no read alignment in at least six consecutive positions) spanned 6,156 bp with Illumina reads, whereas it decreased to 1,787 bp with Nanopore reads. Additionally, assembly regions of coverage zero were much more abundant when aligning Illumina reads (n = 3,624) than when aligning Nanopore reads (n = 54) (fig. 1C). One of these regions is represented in figure 1D, where Nanopore reads uninterruptedly cover this genomic segment with a smooth depth of ∼20×, whereas Illumina reads fail to resolve an intrinsic region where coverage falls to zero, causing the break of contiguity in the assembly. Nanopore reads close Illumina assembly gaps.
In order to assess whether assembly improvement also refines the completeness of coding regions, we first annotated protein-coding genes and noncoding RNA genes. We obtained a 3-fold increase in the recovery of protein-coding genes, noncoding RNA genes and transposable elements from the hybrid assembly in comparison with the Illumina assembly (supplementary table 1, Supplementary Material online). Additionally, we tested completeness by attempting the recovery of conserved single-copy genes from both assemblies. Out of a database containing more than 215 single-copy protozoan orthologs, ∼57% were fully recovered from the hybrid assembly, whereas only ∼29% were recovered from the Illumina assembly. Also, when using a more general database containing over 303 single-copy orthologs conserved across eukaryotic organisms, 68% of these genes were recovered from the hybrid assembly, whereas 48.5% from the Illumina assembly. Together, this demonstrates that Nanopore sequencing helps to mitigate the underestimation of both unique and repetitive coding regions of the genome.
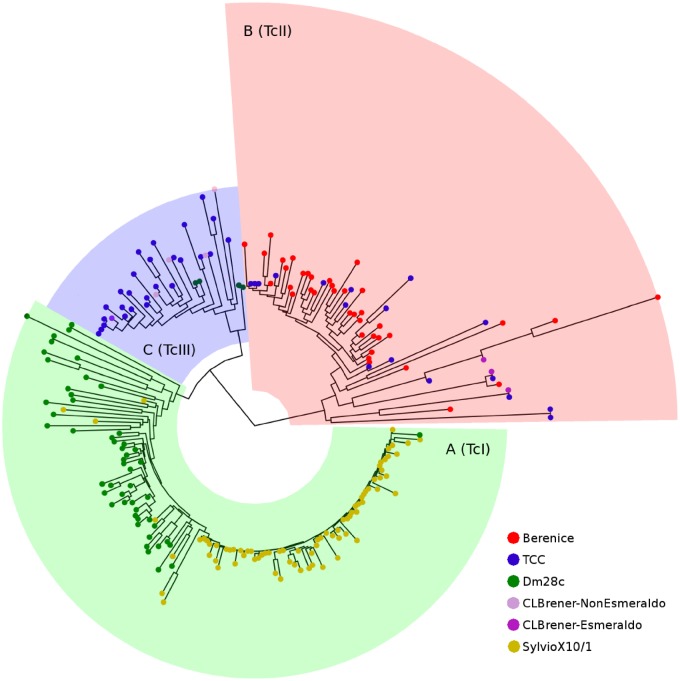
Besides its historical, cultural, and epidemiological relevance for being an isolate from the first clinical case studied by Carlos Chagas, Berenice strain was chosen in order to increase the phylogenetic representativeness of genomes resolved by long-read sequencing, contributing to expand the known genetic diversity of T. cruzi and facilitating the generation of more comprehensive evolutionary inferences. Although initially two groups of T. cruzi were described (I and II) according to biological and biochemical criteria as well as molecular techniques (Tibayrenc et al. 1993), the first study using molecular phylogenetics (based on coding sequences) clearly showed that three major lineages (A, B and C) are present in this parasite (Robello et al. 2000), and the same conclusions were obtained by using new nuclear and mitochondrial sequences (Machado and Ayala 2001). Currently, through a meeting agreement, six groups called “discrete typing units” named TcI–TcVI were proposed, where TcV and TcVI are hybrids of TcII and TcIII (Zingales et al. 2009). In this work, we performed a phylogenetic analysis including Berenice and several available T. cruzi genomes using L1Tc sequences, previously defined as an accurate molecular clock (Berná et al. 2018). The resulting tree clearly shows three major lineages (fig. 2), one comprising sequences from Dm28c and Silvio, belonging to clade A (TcI), other conformed mainly of sequences from TCC and CL-Brener Non-Esmeraldo haplotypes, belonging to clade B (TcIII), and the remaining composed by Berenice, TCC, and CL-Brener Esmeraldo-like haplotypes, belonging to clade C (TcII). Overall, these results show that these new sequencing technologies are finally allowing to solve the complex classification of T. cruzi, strongly confirming the presence of the three major clades A, B and C.
Fig. 2.
—Evolutionary relationships of Trypanosoma cruzi strains. Maximum-likelihood phylogeny constructed with full L1Tc sequences recovered from six T. cruzi genomes.
Here, we used a combination of Illumina and Oxford Nanopore reads to provide the most complete genome assembly of a TcII T. cruzi strain, and constitutes the first application of Nanopore sequencing to resolve a trypanosomatid genome. We compared the assembly continuity and completeness obtained with the most simple library preparation kit of Nanopore with the assembly obtained only using Illumina reads and we obtained a highly improved assembly, similar to the ones obtained using PacBio reads. Even though the coverage and libraries preparation can be optimized, we demonstrate that Oxford Nanopore can be a very valuable technology to improve highly repetitive genomes such as trypanosomatids. This approach has several advantages and can be carried out in every laboratory without any previous training in sequencing, contributing to facilitate the enlargement of genomic resources for protozoan pathogens.
Data Access
Sequencing data generated in this work have been deposited at the NCBI repository under the BioProject accession PRJNA498808.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by Agencia Nacional de Investigación e Innovación (ANII) (DCI-ALA/2011/023–502), “Contrato de apoyo a las políticas de innovación y cohesión territorial,” Fondo para la Convergencia Estructural del Mercado Común del Sur (FOCEM) 03/11; UK Research and Innovation via the Global Challenges Research Fund under grant agreement “A Global Network for Neglected Tropical Diseases” grant number MR/P027989/1; and Fundação de Amparo à Pesquisa do Estado de Minas Gerais (CBB-AUC-00030-15). S.P., G.I., G.G., and C.R. are members of the “Sistema Nacional de Investigadores (ANII)”; F.D.-V. has an ANII doctoral fellowship no. POS_NAC_2016_1_129916. The authors declared they have no conflicts of interest.
Author Contributions
C.R. and G.I. conceived the idea. R.C.M. processed and prepared samples. F.D.-V. and G.G. prepared libraries and performed Illumina and Nanopore sequencing. F.D.-V., G.I., and S.P. analyzed the data. F.D.-V., G.I., S.P., and C.R. wrote the manuscript. All authors approved the final version of the manuscript.
Data deposition: This project has been deposited at the NCBI under the accession PRJNA498808.
Literature Cited
- Belyeu JR, et al. 2018. SV-plaudit: a cloud-based framework for manually curating thousands of structural variants. Gigascience 7:giy064.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27(2):573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berná L, et al. 2018. Expanding an expanded genome: long-read sequencing of Trypanosoma cruzi. Microb Genom. 4:e000177.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callejas-Hernández F, Rastrojo A, Poveda C, Gironès N, Fresno M.. 2018. Genomic assemblies of newly sequenced Trypanosoma cruzi strains reveal new genomic expansion and greater complexity. Sci Rep. 8(1):14631.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagas C. 1909. Nova tripanozomiaze humana: estudos sobre a morfolojia e o ciclo evolutivo do Schizotrypanum cruzi n. gen., n. sp., ajente etiolojico de nova entidade morbida do homem. Mem Inst Oswaldo Cruz 1(2):159–218. [Google Scholar]
- de Lana M, et al. 1996. Characterization of two isolates of Trypanosoma cruzi obtained from the patient Berenice, the first human case of Chagas’ disease described by Carlos Chagas in 1909. Parasitol Res. 82(3):257–260. [DOI] [PubMed] [Google Scholar]
- Deane LM. 1964. Animal reservoirs of Trypanosoma cruzi in Brazil. Rev Bras Malariol Doencas Trop. 16:27–48. [PubMed] [Google Scholar]
- El-Sayed NM, Myler PJ, Bartholomeu DC, et al. 2005. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science 309(5733):409–415. [DOI] [PubMed] [Google Scholar]
- El-Sayed NM, Myler PJ, Blandin G, et al. 2005. Comparative genomics of trypanosomatid parasitic protozoa. Science 309(5733):404–409. [DOI] [PubMed] [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate Maximum-Likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. [DOI] [PubMed] [Google Scholar]
- Kalvari I, et al. 2018. Non-coding RNA analysis using the Rfam database. Curr Protoc Bioinformatics 62(1):e51. 10.1002/cpbi.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34(18):3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Chan PP.. 2016. tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado CA, Ayala FJ.. 2001. Nucleotide sequences provide evidence of genetic exchange among distantly related lineages of Trypanosoma cruzi. Proc Natl Acad Sci U S A. 98(13):7396–7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR.. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29(22):2933–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, et al. 2015. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 43(Database issue):D130–D137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pita S, Díaz-Viraqué F, Iraola G, Robello C.. 2019. The Tritryps comparative repeatome: insights on repetitive element evolution in Trypanosomatid pathogens. Genome Biol Evol. 11(2):546–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassi A, Rassi A, Marin-Neto JA.. 2010. Chagas disease. Lancet 375(9723):1388–1402. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A.. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16(6):276–277. [DOI] [PubMed] [Google Scholar]
- Robello C, Gamarro F, Castanys S, Alvarez-Valin F.. 2000. Evolutionary relationships in Trypanosoma cruzi: molecular phylogenetics supports the existence of a new major lineage of strains. Gene 246(1-2):331–338. [DOI] [PubMed] [Google Scholar]
- Salgado JA, Garcez PN, de Oliveira CA, Galizzi J.. 1962. Revisão clínica atual do primeiro caso humano descrito da doença de Chagas. Rev Inst Med Trop Sao Paulo 4:330–337. [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Tan MH, et al. 2018. Finding Nemo: hybrid assembly with Oxford Nanopore and Illumina reads greatly improves the clownfish (Amphiprion ocellaris) genome assembly. Gigascience 7:gix137.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibayrenc M, et al. 1993. Genetic characterization of six parasitic protozoa: between random-primer DNA typing and multilocus enzyme electrophoresis. Proc Nac Acad Sci USA 4:1335–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. 2017. Chagas disease (American Trypanosomiasis). Available from: http://www.who.int/mediacentre/factsheets/fs340/en/
- Wick RR, Judd LM, Gorrie CL, Holt KE.. 2017. Completing bacterial genome assemblies with multiplex MinION sequencing. Microb Genom. 3: e000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin AV, et al. 2013. The MaSuRCA genome assembler. Bioinformatics 29(21):2669–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin AV, et al. 2017. Hybrid assembly of the large and highly repetitive genome of Aegilops tauschii, a progenitor of bread wheat, with the MaSuRCA mega-reads algorithm. Genome Res. 27(5):787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingales B, et al. 2009. A new consensus for Trypanosoma cruzi intraspecific nomenclature: second revision meeting recommends TcI to TcVI. Mem Inst Oswaldo Cruz 104(7):1051–1054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.