Figure 1.
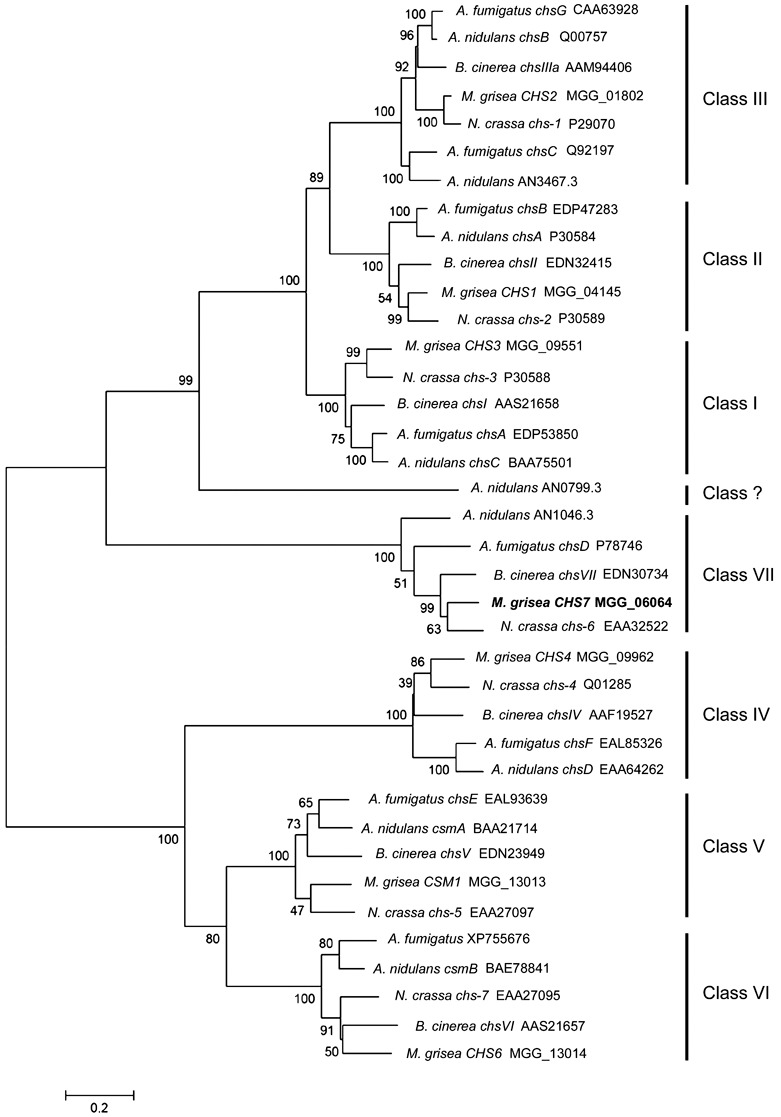
Phylogenetic tree based on alignment of the predicted Magnaporthe grisea CHS7 gene product with the predicted gene products of known and predicted chitin synthase (CS)‐encoding genes. Tree reliability was tested using bootstrap resampling with 1000 replications. Together with gene names, where assigned, the GenBank or Swissprot accession numbers are given, or, alternatively, and for all the Magnaporthe predicted proteins used, the Broad Institute database ID number is given. Note that AN0799.3 (marked Class ?) seems to be a divergent CS which, although it has characteristics of the class I, II and III CSs [PFAM domains PF1644 (chitin synthase 1) and PF08407 (chitin synthase 1 N‐terminal domain)], lacks the close similarity that the other members of these classes exhibit. This protein shares only 28% of its amino acid sequence with Aspergillus nidulans ChsC protein (class I). For comparison, the A. nidulans ChsA (class II) and ChsC proteins have 48% of amino acids in common, a value more typical between the class I and II CSs, whereas, within the various CS classes, typically 60%–70% of amino acids are conserved between species. Numbers on the branches indicate the bootstrap value for each branch and the bar indicates 0.2 distance units.
