Figure 3.
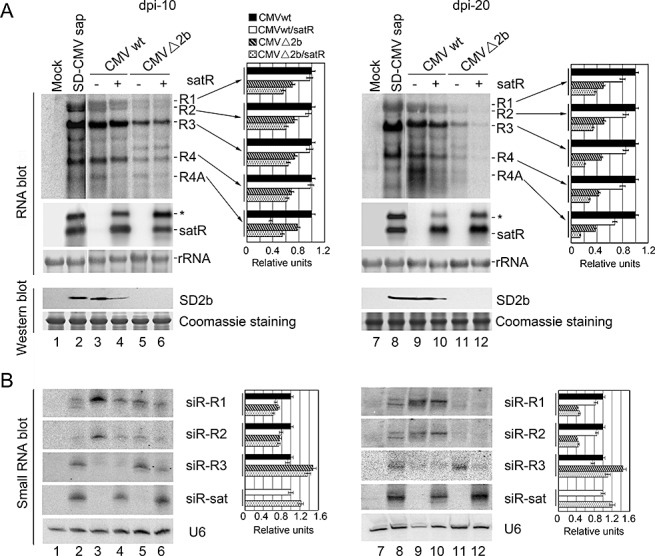
RNA gel blot analysis of viral RNAs, satellite RNA (satRNA) and pathogen‐derived small interfering RNA (siRNA) accumulations in systemically infected leaves of Agrobacterium‐inoculated Nicotiana benthamiana. (A) Cucumber mosaic virus (CMV) RNAs and satRNA accumulation were examined in similar layers of systemically infected leaves of Agrobacterium‐inoculated N. benthamiana, as described in Fig. 2, collected at 10 days post‐inoculation (dpi) (left panel) and 20 dpi (right panel). Pools of seven plants were analysed for each inoculation. Hybridization was performed with 32P‐labelled SD‐CMV genomic RNA 3′‐UTR‐ and satRNA‐specific probes, respectively. Six micrograms of total RNAs were loaded. Methylene blue‐stained ribosome rRNA was used as loading control. Accumulation of the 2b protein was also detected by protein immunoblot with anti‐2b polyclonal antiserum. ‘Coomassie’ staining was used as loading control. (B) CMV RNAs‐ and satRNA‐derived siRNAs were examined in the leaf samples, as described in (A). 32P‐labelled in vitro transcripts from viral RNA1, RNA2, RNA3 and satRNA were used as probes. U6 RNA hybridizations are shown as loading control. Quantifications of each CMV RNA and siRNA relative to loading control RNA are shown on the right of each panel using ImageQuant TL (GE Healthcare Life Sciences). RNAs obtained from CMVwt infection were arbitrarily assigned a value of 1.0. An asterisk denotes possible double‐stranded satRNA.
