Figure 4.
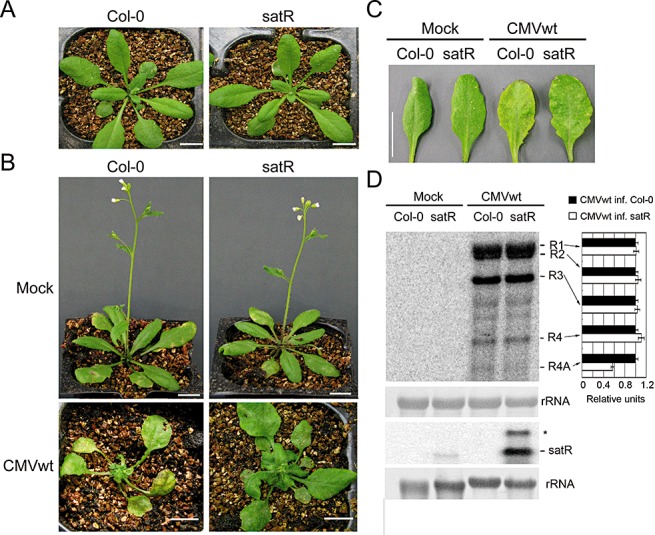
Analysis of effect of ShanDong satellite RNA (SD‐satRNA) on pathogenicity in Arabidopsis. (A) The morphological phenotype of 35S‐satR‐transgenic plants (satR) was comparable with that of wild‐type (WT) Arabidopsis (Col‐0). (B, C) Col‐0 and SD‐satR plants were inoculated with CMVwt sap, extracted from Nicotiana benthamiana leaves infected with CMVwt via Agrobacterium infection. Mock inoculation was used as a control. The disease symptoms of whole plants (B) and early systemically infected leaves (C) were photographed at 12 days post‐inoculation (dpi). Scale bar, 1 cm. (D) RNA gel blot analysis was performed to detect viral RNAs and satRNA accumulation in systemically noninoculated leaves at 12 dpi. Methylene blue‐stained rRNA served as loading control. Quantifications of each CMV RNA relative to loading control RNA are shown on the right of the panel using ImageQuant TL (GE Healthcare Life Sciences). RNAs obtained from CMVwt‐infected Col‐0 were arbitrarily assigned a value of 1.0. An asterisk denotes possible double‐stranded satRNA.
